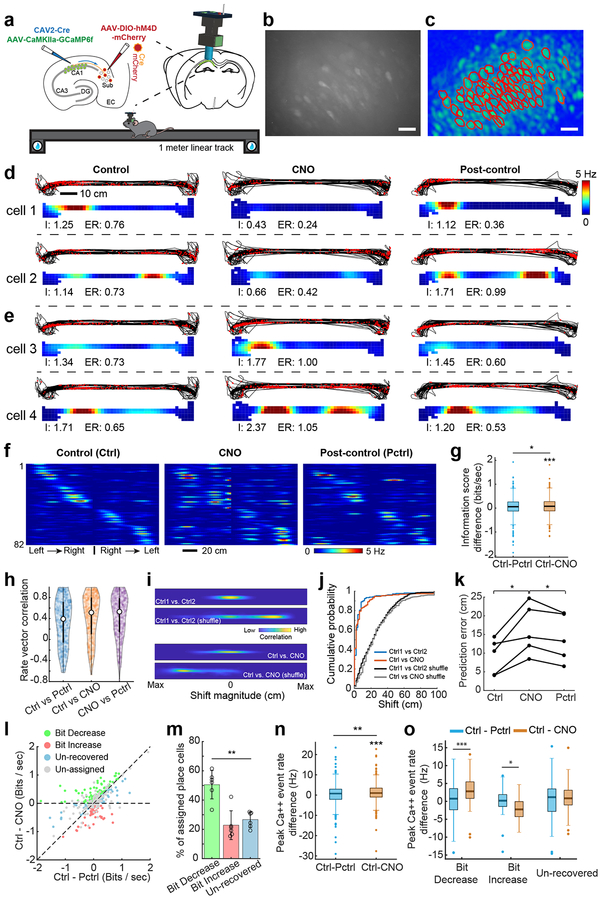
Fig. 5. CA1-projecting SUB neurons modulate place-specific activity of CA1 neurons in the linear track space.
Experiments described in the figure were independently repeated in 6 CNO-treated mice, each with similar results obtained. For control data, experiments were independently repeated in 3 mice, each with similar results obtained. a, Top left, a schematic illustration of AAV1 injection for targeted expression of GCaMP6f in CA1 excitatory neurons, and a second dual injection of CAV2-Cre in CA1 and AAV2-DIO-hM4D in SUB for targeted expression of DREADDs in the CA1-projecting SUB excitatory neurons. Top right, a schematic depicts a miniaturized fluorescent microscope (miniscope) used to image in vivo calcium signals in CA1 neurons in awake behaving mice. The implanted GRIN lens (shown in blue) and fixed miniscope baseplate allow reliable, repeated imaging of the same group of neurons over 2 weeks. The lower cartoon depicts a mouse running on a 1-meter linear track during in vivo calcium imaging. Water rewards are placed on both ends of the linear track. b, Representative maximum intensity projected image showing recorded CA1 neurons from three combined 15 min imaging sessions (control, CNO, post-control) across days. The calcium imaging videos were motion corrected and aligned across sessions, scale bar = 25 μm. c, A spatial footprint profile image shows extracted neurons (red contours) using the CNMF-E algorithm (see the Methods for details) based on the combined video in b, scale bar = 25 μm. d, Two sets of panels display tracking data (black lines) with superimposed red dots depicting sites where Ca++ events occurred (upper) and corresponding calcium activity rates (lower) for two example cells (cell 1 and cell 2) from the bit-decrease group (see classification below) on the linear track. Each bottom panel is a color-coded rate map showing the averaged spatial distribution of calcium event rates of the same CA1 neuron mapped to animal position on the linear track. Each line shows the event plot and rate maps of control (left), CNO (center), and post-control (right) sessions from the same CA1 place cell over about 2 weeks (2–3 days between sessions). “I” indicates the spatial information score (bits/second), “ER” indicates the mean calcium event rate. e, Same organization as d, these panels show the activities of two example cells (cell 3 and cell 4) from the bit-increase (see classification below) group. f, A color coded population event rate map organized by spatial position for left to right track traversals and then right to left traversals of all place cells from one representative mouse during control (Ctrl, left), CNO (center), and post-control (Pctrl, right) sessions. The depicted cell order is unchanged across the sessions of control, CNO and post-control as determined initially in the control session. Each line shows activity of one place cell. Color indicates event rate (scale bar). The overall rate map correlation values (Pearson’s) between control and post control sessions, and between control and CNO sessions are, respectively, 0.61 and 0.54. g, Comparison of the difference of spatial information scores (bits/second) between Ctrl and Pctrl (Ctrl –Pctrl, two-tailed t-test against zero: p = 0.15), and between Ctrl and CNO (Ctrl – CNO, two-tailed t-test against zero, p = 1.3 × 10−4) across all 347 place cells recorded from 6 mice. Differences between Ctrl – Pctrl and Ctrl – CNO are also observed (two-tailed, paired t-test, p = 0.03). h, A violin plot showing the distribution of individual place cell’s rate vector correlation coefficients (Pearson’s) for all session combinations (“control versus CNO”, “control versus post-control”, and “CNO versus post-control”). The median value for Ctrl vs Pctrl, Ctrl vs CNO, and CNO vs PCtrl are 0.40, 0.51, and 0.53 (n = 347 place cells from 6 mice). The white points indicate median values, and thin black lines extend to the most extreme values within 1.5 times of the interquartile range of the median. The filled color width represents a density estimate of the distribution of values along the y axis. i, Spatial cross-correlograms of example CA1 cells from the saline-treated experiment (Ctrl1 vs Ctrl2) and CNO-treated experiment (Ctrl vs CNO). Shuffled examples were obtained by randomly pairing rate maps across the same comparison sessions as for the example cells. j, The distributions of correlation-peak shift magnitudes for the place cells in the saline experiment (blue line) and the CNO experiment (red line) differ significantly from the corresponding shuffled distributions (p = 3.17 × 10−64, two-tailed, two-sample Kolmogorov–Smirnov test (KS), n = 174 cells from Ctrl1 vs Ctrl2 and 1000 shuffles from Ctrl1 vs Ctrl2; p = 1.48 × 10−101, two-sample KS, two-tailed, n = 347 cell from Ctrl vs CNO and 1000 shuffles from Ctrl vs CNO). Shuffled distributions were obtained by randomly pairing place maps 1,000 times across the indicated sessions. There is no significant difference between the distribution of Ctrl vs CNO (red line) and the Ctrl1 vs Ctrl2 (blue line) (p = 0.38, two-sample KS, two-tailed). k, Quantification of the prediction errors between predicted trajectories and actual trajectories for decoding accuracy using the trained model based on the first control session, which supports the observations in Supplementary Fig. 8a. Each line represents the prediction errors of Ctrl, CNO and Pctrl sessions from one mouse. Significantly higher prediction errors are observed in CNO sessions compared to those in Ctrl (p = 0.016, two-tailed, paired t-test) and Pctrl (p = 0.015, two-tailed, paired t-test) sessions. n = 5 mice. l, Recorded CA1 place cells can be classified into 3 non-overlapping groups termed bit-decrease, bit-increase, and un-recovered (see Methods for more information about the group classification), based on the statistical significance of differences in information scores (bit/sec) between CNO and Ctrl, and between CNO and Pctrl. Statistical testing employed a jackknife resampling method for each place cell with appropriate corrections for error terms. Un-assigned place cells did not pass the statistical test and were excluded from further categorization analysis. On the scatter plot, the x-axis is Ctrl – Pctrl (the difference of spatial information scores between Ctrl and Post-ctrl) and the y-axis is Ctrl – CNO (the difference of information scores between Ctrl and CNO). m, Of the 201 place cells that show significant differences (assigned place cells) from 6 mice, 50% show decreased information scores in CNO sessions compared to the control and post-control sessions (bit-decrease group, green bar). A smaller subset (~ 23%) show increased information scores in CNO compared to the control and post-control (bit-increase group, red bar). The remaining ones are the unrecovered group which accounts for ~ 27% of place cells. Comparing the mean percentages of each type seen in each mouse, a significant difference in the % of place cells among these three groups is observed (p = 0.002, repeated measures ANOVA, n = 6 mice). Data are presented as mean ± SE in the bar plot. n, Comparison of the difference of peak calcium event rates between Ctrl and Pctrl (Ctrl –Pctrl, two-tailed t-test against zero, p = 0.32), and between Ctrl and CNO (Ctrl –CNO, two-tailed t-test against zero, p = 1.6 × 10−5) across all 347 place cells recorded from 6 mice. Differences between Ctrl – Pctrl and Ctrl – CNO are also observed (two-tailed, paired t-test, p = 0.004). o, Comparisons of peak calcium event rates between Ctrl – Pctrl and Ctrl – CNO in bit decrease (two-tailed, paired t-test, p = 3 × 10−7, n = 97 cells), bit increase (two-tailed, paired t-test, p = 0.027, n = 48 cells) and un-recovered groups (two-tailed, paired t-test, p = 0.20, n = 56 cells), respectively. For the box plots throughout the figure, the three box lines from top to bottom represent the 25th, 50th (median), and 75th percentile of data values of the samples. The whiskers extend to the most extreme values within 1.5 times of the interquartile range of the median.