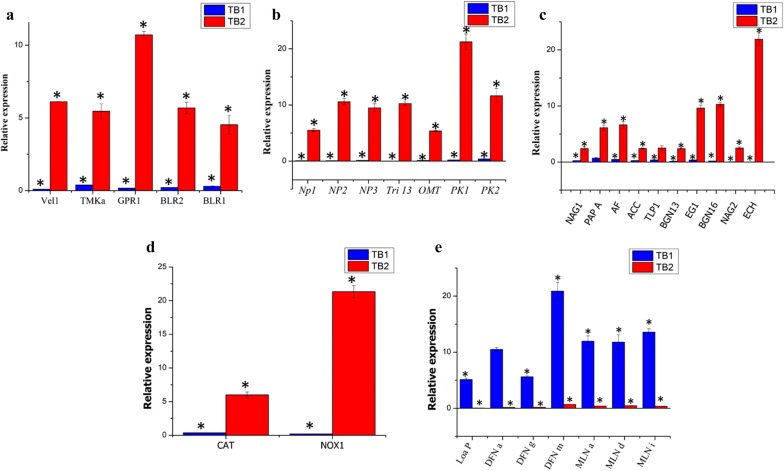
Fig. 1.
Influence of simultaneous (TB1) and sequential (TB2) inoculation based co-cultivation on the gene expression of (a) the T. asperellum morphology related genes [Velvet (Vel1), mitogen-activated protein Kinase (TMKa), G protein receptor 1 (GPR1), blue-light-regulated genes (BLR1 and BLR2) and ENVOY (ENV1)], (b) secondary metabolism related genes [non-ribosomal peptide synthetase (NP1 and NP2), Putative ferrichrome synthetase (NP3), Cytochrome P450 (Tri 13) 1,O-methyl transferase (OMT) and Polyketide synthetase (PK1 and PK2)]; (c) mycoparasitism-related genes [chitinase (ECH), β-1,3-glucanase (BGN13), β-1,6-glucanase (BGN16), β-1,4-glucanase (Egl), N-acetyl-glucosaminidases (NAG1 and NAG2), aspartyl protease (PAP A), trypsin-like protease (TLP 1) and α-l-arabinofuranosidases (AF)]; and plant growth promoting enzyme [1-Aminocyclopropane-1-carboxylate deaminase (ACC)]. d Anti-oxidant genes [NADPH oxidase (NOX), catalase (CAT)] and (e) genes encoding B. amyloliquefaciens macrolactin and difficidin in the co-culture [intrinsic terminators located within the polyketide synthase (PKS) gene cluster encoding for the antibiotic difficidin (DFN) (Loa P), beginning, middle and end of the difficidin operon (DFN A, DFN G, and DFN M); beginning, middle and end of the macrolactin operon (MLN a, MLN d, and MLN i)]. Bars represent the standard error of the mean values of three biological replicates. ∗ represent significant differences between the axenic and co-culture (P < 0.05)