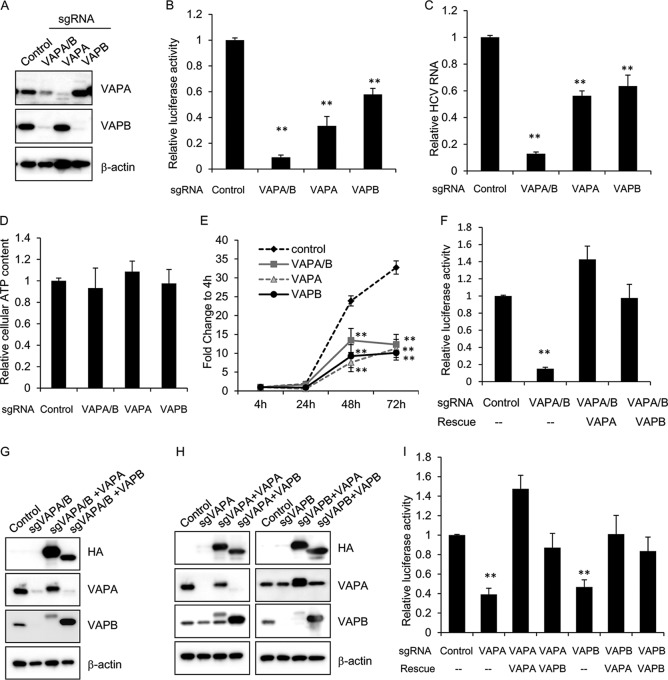
FIG 1.
VAPA and VAPB are required for efficient HCV replication. (A) Huh7.5.1 cells were stably transduced with lentivirus encoding a puromycin resistance marker, Cas9, and a single guide RNA (sgRNA) targeting the indicated gene(s). Lysates from the stable knockout cell pools were then immunoblotted for the indicated proteins. (B) Huh7.5.1 knockout cell pools as described in panel A were infected with NanoLuc-HCV for 3 days, and luciferase activity was measured. Values are means ± standard errors of the means of three independent experiments and are normalized to the level of the control. **, P < 0.001, for results compared to the result with the control. (C) Huh7.5.1 knockout cell pools as described in panel A were infected with JFH-1 for 3 days before viral RNA was quantified by quantitative reverse transcription-PCR. Values are normalized to the result with the control. **, P < 0.001, for results compared to the result with the control. (D) Cell viability of Huh7.5.1 cells stably transduced with lentivirus encoding sgRNA targeting the indicated gene was measured by CellTiter-Glo. Values are normalized to the level of the control. (E) Huh7.5.1 knockout cell pools as described in panel A were transfected with a subgenomic replicon encoding a Renilla luciferase reporter. Luciferase activity was measured at 4, 24, 48, and 72 h posttransfection, and data are plotted as fold change over values at 4 h to control for transfection and initial translation. **, P < 0.001, for results compared to the result with the control. (F) VAPA/B double knockout Huh7.5.1 cell pools were transduced with the indicated lentiviral vectors to express HA-tagged sgRNA-resistant VAPA or VAPB and infected with NanoLuc-HCV for 3 days, and luciferase activity was measured. Values are means ± standard errors of the means of three independent experiments and are normalized to the level of the control. **, P < 0.001, for results compared to the result with the control. (G) Stably transduced cells as described in panel F were immunoblotted for the indicated proteins. sgVAPA/B, sgRNA targeting VAPA/B. (H) Indicated knockout cell pools were transduced with the indicated lentiviral vectors to express HA-tagged VAPA or VAPB and immunoblotted for the indicated proteins. (I) Stably transduced cells as described in panel H were infected with NanoLuc-HCV for 3 days, and luciferase activity was measured. Values are means ± standard errors of the means of three independent experiments and are normalized to the level of the control.