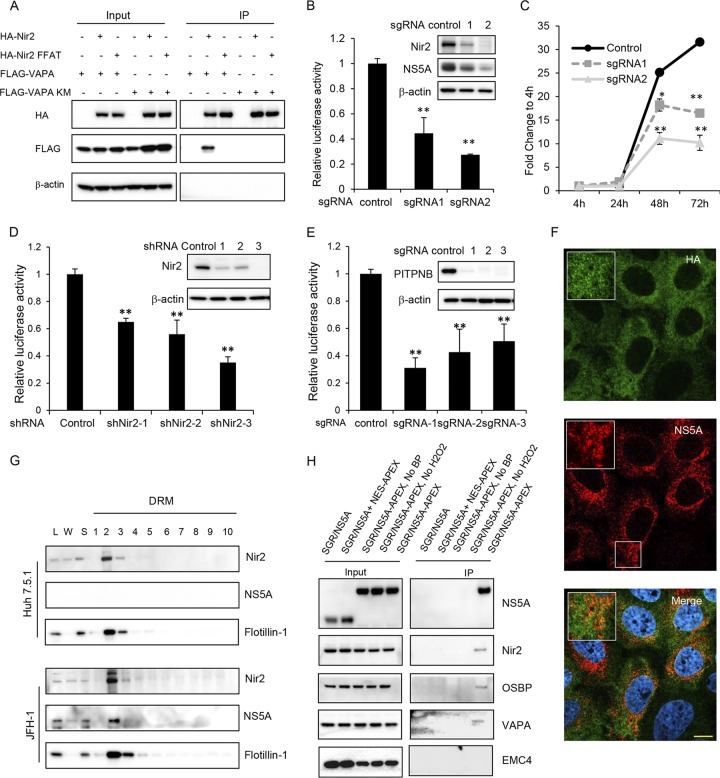
FIG 5.
Nir2 is required for HCV replication and is in close proximity to HCV ROs. (A) Coimmunoprecipitation of VAPA with Nir2 protein. 293T cells were cotransfected to express FLAG-tagged Nir2 or the Nir2 FFAT mutant with HA-tagged VAPA or the VAPA KM→DD mutant. Forty-eight hours later, cells were lysed and immunoprecipitated with HA antibody, followed by immunoblotting with the indicated antibodies. (B) Huh7.5.1 cells stably expressing a negative control or two independent Nir2 sgRNAs with Cas9 were infected with NanoLuc-HCV for 3 days before luciferase activity was measured. Values are normalized to the level of the control. **, P < 0.001, for results compared to the result with the control. Inset, Huh7.5.1 cell pools as described above were infected with the JFH-1 strain of HCV followed by immunoblotting for the indicated proteins. (C) Huh7.5.1 cell pools described as in panel B were transfected with a subgenomic replicon encoding a Renilla luciferase reporter. Luciferase activity was measured at 4, 24, 48, and 72 h posttransfection, and data are plotted as fold change to values at 4 h to control for transfection and RNA translation. *, P = 0.01; **, P < 0.005, for results compared to the result with the control. (D) OR6 replicon cells were transduced with the indicated shRNAs, and replication was assessed at 72 h postransduction by Renilla luciferase activity. Values are normalized to the level of the control. **, P < 0.001, for results compared to the result with the control. Inset, OR6 cell pools as described were immunoblotted for the indicated proteins. (E) Huh7.5.1 cell pools stably expressing a negative control or three independent PITPNB sgRNAs with Cas9 were infected with NanoLuc-HCV for 3 days before luciferase activity was measured. Values are normalized to the level of the control. **, P < 0.001, for results compared to the result with the control. Inset, Huh7.5.1 cell pools as described were immunoblotted for the indicated proteins. (F) Nir2 knockout Huh7.5.1 cell pools stably overexpressing HA-tagged Nir2 were infected with JFH-1. Three days later, cells were fixed and stained with antibodies against NS5A (green) and HA (red) with DAPI nuclear counterstaining (blue). Bar, 10 μm. (G) Uninfected or JFH-1 infected Huh7.5.1 cells were prepared as in the legend of Fig. 3C, and detergent-resistant membranes (DRMs) were fractionated on an iodixanol density gradient. Fractions were analyzed by SDS-PAGE, followed by immunoblotting for the indicated proteins. Fractions are numbered from 1 to 10 in order from top to bottom (light to heavy). L, cell homogenate; W, water-soluble supernatant. (H) Nir2 is in close proximity to NS5A. Huh7.5.1 stably expressing a subgenomic replicon (SGR/NS5A) or coexpressing the SGR/NS5A replicon and NES-APEX2 or a subgenomic replicon with APEX2-tagged NS5A (SGR/NS5A-APEX) were treated, where indicated, with biotin-phenol (BP)-containing medium for 30 min, followed by hydrogen peroxide for 1 min to biotinylate APEX2-proximal proteins prior to quenching and lysis. Biotinylated proteins were affinity isolated with streptavidin beads, and lysates were resolved by SDS-PAGE. Immunoblotting was performed for the indicated proteins for both input and immunoprecipitated (IP) samples.