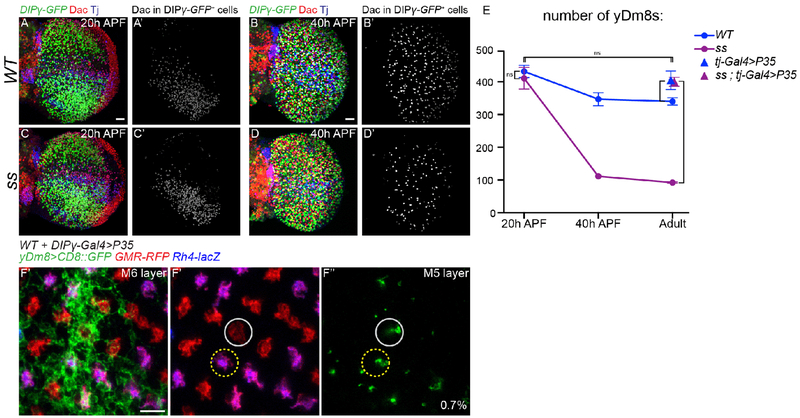
Fig. 4. Apoptosis of excess Dm8s ensures the numerical matching of R7s and Dm8s:
(A-D) DIPγ-GFP expression in the optic lobe in WT (A,B) and ss mutant (C,D) at 20 hours (A and C) and 40 hours After Pupa Formation (APF) (B and D). yDm8s were labelled by segmenting the Dac staining from the GFP staining (A’-D’, in grey). Scale bar: 20 μm in A for (A-D). (E) Number of yDm8s per optic lobe (DIPγ-GFP+,Dac+ cells, n=4-6 optic lobes per genotype). Bars show the mean +/− SD. ****p<0.0001, one-way ANOVA, Tukey test. (F) Proximodistal view of yDm8s labelled with CD8::GFP in WT upon cell death inhibition by mis-expressing P35 in DIPγ expressing neurons. A single yDm8 mis-paired with a pR7 (circled in grey) showed by dense GFP staining at the level of the M6 layer (E’) and its home column at the level of the M5 layer (F” and F”’) (mis-paired yDm8s=0.7%, n=428). Scale bar: 5 μm.