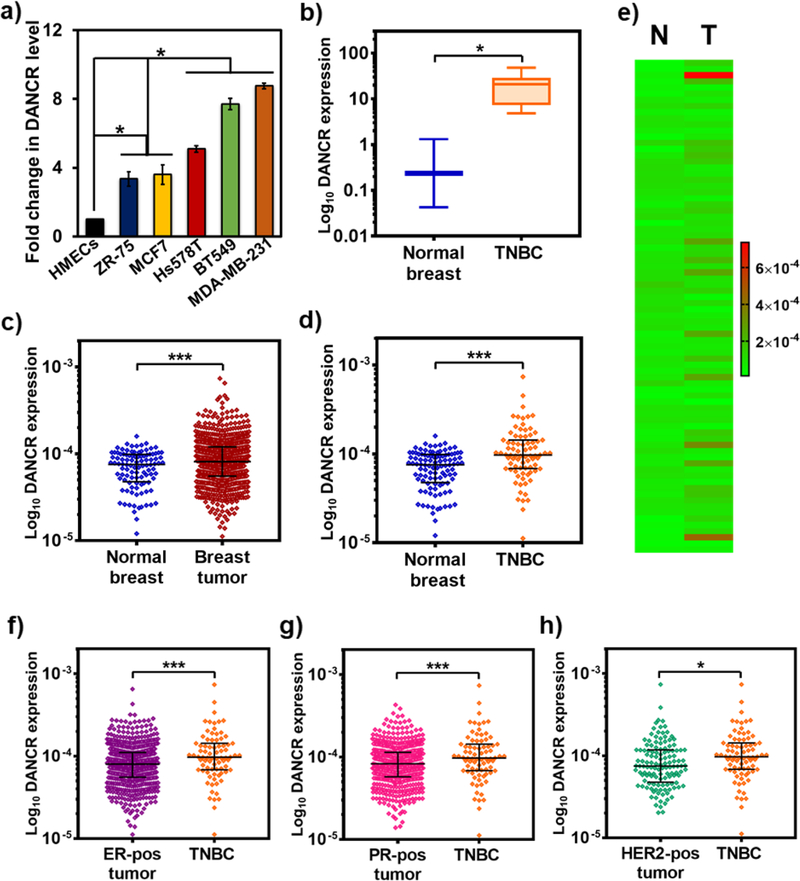
Figure 2.
DANCR is significantly overexpressed in TNBC. (a) Endogenous expression of DANCR is significantly elevated in all breast cancer cell lines: MCF7 (luminal A; ER+, PR+/−, Her2−), ZR-75 (luminal B, ER+, PR+/−, Her2+), and Hs578T, BT549, and MDA-MB-231 (claudin-low ER−, PR−, Her2−), compared to HMECs, and even more so in the TNBC cell lines. (b) Quantification of DANCR expression from a breast cancer array containing cDNA from normal subjects (n=4) and breast cancer patients shows that DANCR is highly upregulated in tumor samples from TNBC patients (n=12), in comparison with breast tissues from normal subjects. (c) Differential gene expression analysis for DANCR performed for 104 normal and 790 breast cancer samples from TCGA database shows a significant upregulation of DANCR in breast cancer samples, particularly in 80 TNBC samples (d), compared to normal levels. (e) Heat map comparing and demonstrating the heterogeneity of DANCR expression in the individual normal (N) and TNBC (T) samples from TCGA database. DANCR expression is significantly higher in TNBC samples than breast cancer samples expressing (f) ER (n=584), (g) PR (n=508), and (h) HER2 (n=128) receptors (error bars denote s.e.m., *p<0.05, ***p<0.0005).