Fig. 2 |. Comparison of representative inverting and retaining catalytic mechanisms and enzyme topologies for substrate interactions.
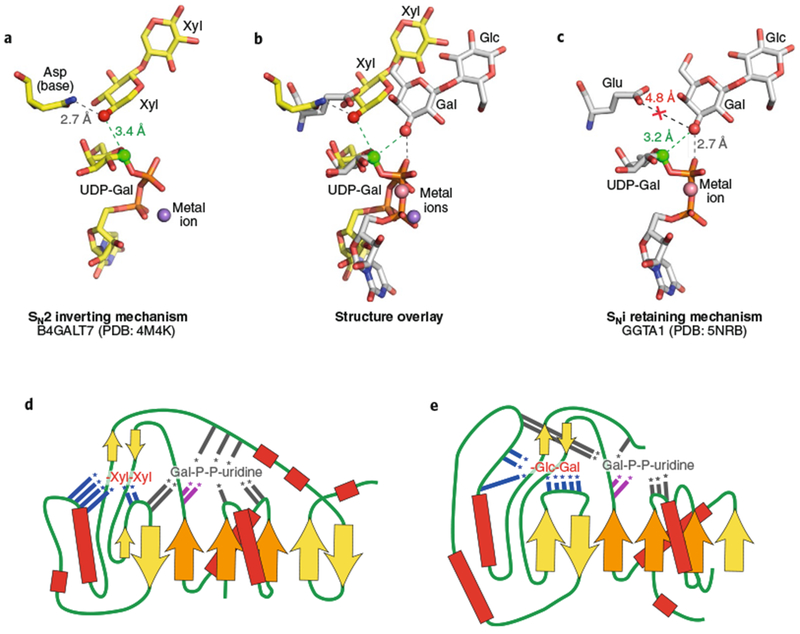
Bound ligands for B4GALT7 (a, PDB: 4M4K26) and GGTA1 (c, PDB: 5NRD25) are displayed along with putative catalytic bases. a, The inverting SN2 catalytic mechanism for B4GALT7 is catalyzed by deprotonation of the Xyl acceptor hydroxyl (red sphere) by the catalytic base (Asp211 in the wild type enzyme mutated to Asn211 in PDB ID 4M4K) and attack of the anomeric carbon (green sphere) in line with the departing nucleotide leaving group. c, GGTA1 has a retaining SNi-like mechanism in which the nucleophile position (red sphere) is shifted relative to the inverting enzymes. The equivalent catalytic base residue (Glu317) is too far away for deprotonation, but the acceptor Gal hydroxyl (red sphere) is positioned adjacent to the β-phosphate oxygen for deprotonation (gray dotted line) and in proximity to attack the anomeric carbon (green sphere). b, Overlay of the two structures based on the Gal C1-phosphate oxygen bond illustrates the differences in geometry of nucleophilic attack on the anomeric carbons. d,e, Donor and acceptor binding sites for B4GALT7 (d) and GGTA1 (e) are displayed as topology diagrams illustrating the positions of residues that interact with donors and acceptors in loops extending from the core β-sheets of the Rossmann fold. β-Sheets are indicated as filled arrows (orange for conserved Rossmann-fold elements and yellow for nonconserved sheets among GT-A fold enzymes) and helices as red boxes connected by loop regions (green lines). Positions of interacting residues are indicated by colored lines with asterisks (gray, interacting with donors; blue, interacting with acceptors; magenta, DxD motif). Acceptors for each enzyme (B4GALT7: Xyl-β-1,4-Xyl and GGTA1: Gal-β-1,4-Glc) are indicated in abbreviated form, but with inverted orientation from standard nomenclature to indicate the proximity of the terminal sugar acting as nucleophile in the glycosyltransfer reaction.
