Figure 2.
Establishment of a Processing Protocol for Quantitative Measurement of Tumor Volume
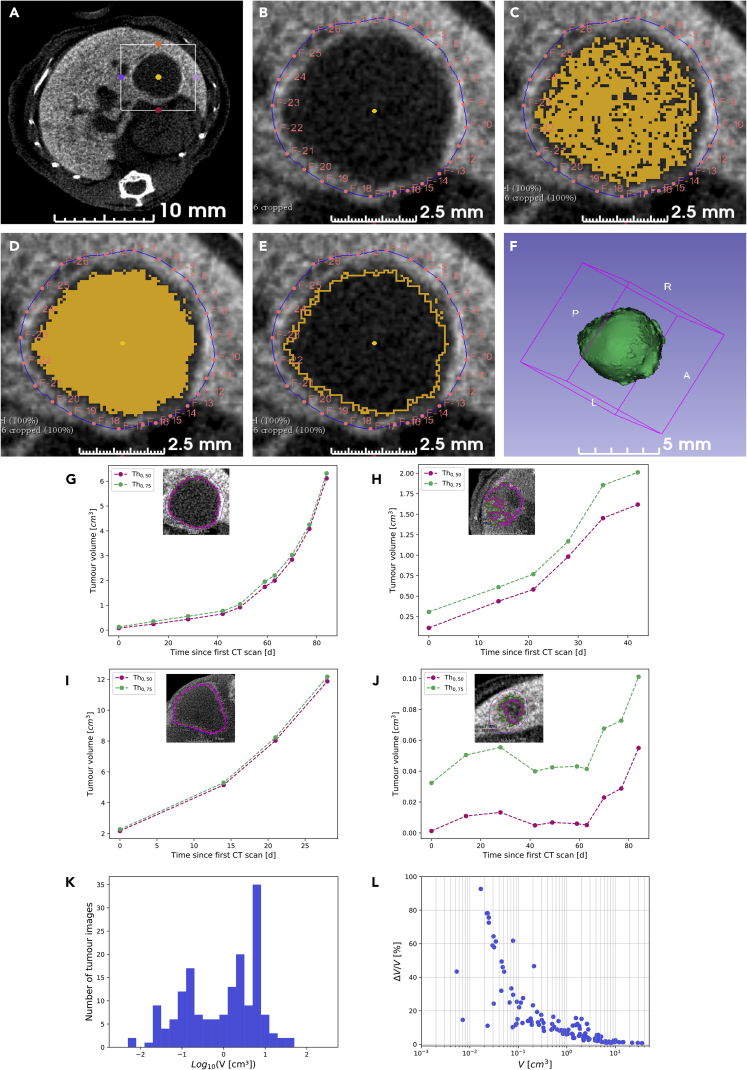
(A–F) Images exemplifying the semiautomatic segmentation protocol we followed for tumor volume measurements. The region of interest enclosing the tumor to analyze for quantifications (A) was processed with manual demarcation of the area of segmentation by means of red fiducial markers (B) and application of the predefined contrast threshold to select voxels with a value inferior to the contrast threshold (C). Morphological smoothing to connect the area of voxels was then applied (D), and segmented area was defined according to the predefined contrast threshold (E). Images corresponding to the whole tumor were processed, and values were merged for a 3D reconstruction of the tumor volume (F).
(G–J) Graphs reporting representative examples of longitudinal follow-up of tumor volume growth, corresponding to the largest tumor reported in Figure 1B (G; tumor M1-T4), a medium tumor (H; tumor M5-T1), a big tumor (I; tumor M4-T1), and a small tumor (J; tumor M1-T3). Tumor volume measurements are shown for the two defined contrast thresholds, Th0.50 (pink) and Th0.75 (green). Images shown in each panel correspond to transverse slices at the tumor maximal diameter acquired at D49 (G and J), D21 (H), and D15 (I); the two-contrast thresholds applied are reported on images.
(K) Histogram of the overall measured tumor volumes, defined as V = (V0.50 + V0.75)/2.
(L) Graph reporting the tumor volume uncertainty, defined as ΔV = (V0.75 - V0.50)/2. Note that major uncertainties were observed mainly for tumors with a volume less than 10−1 cm3.
See also Figure S3.