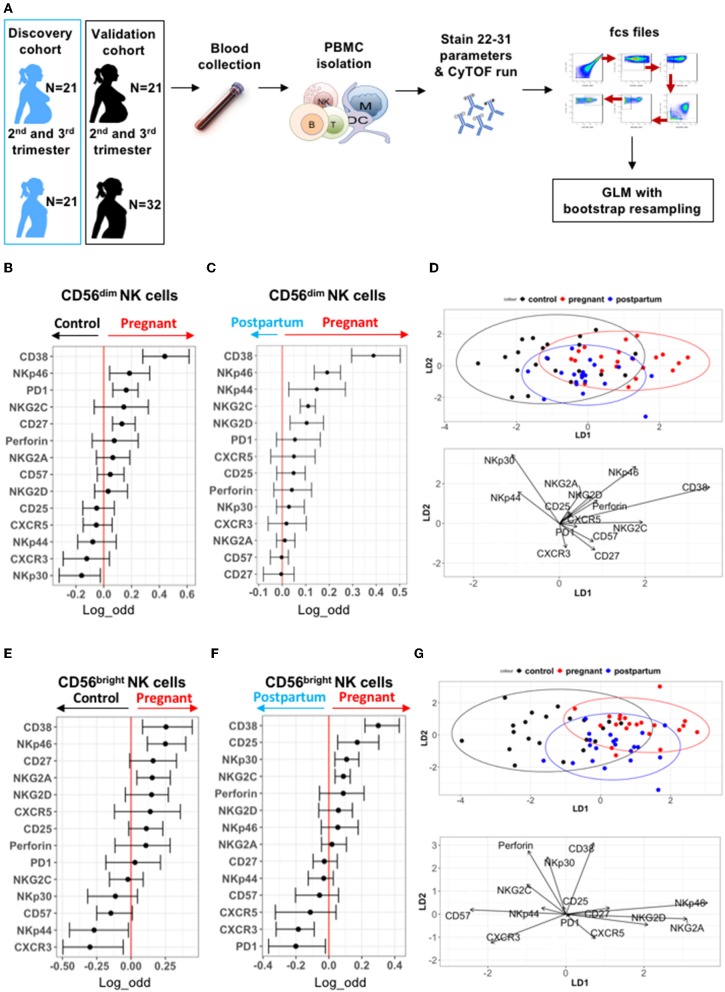
Figure 2.
Deep profiling of CD56dim and CD56bright NK cells in non-pregnant and pregnant women from discovery cohort. (A) PBMCs from controls (N = 21), pregnant women (N = 21), and post-partum women (N = 21) in discovery cohort were isolated from blood samples. PBMCs were isolated and stained for mass cytometry. Data obtained were analyzed using a linear regression model, GLM. (B,C) Markers predictive of CD56dim NK cells in control vs. pregnant women in discovery cohort were assessed by GLM with bootstrap resampling. The markers are listed on the y-axis and the x-axis represents the log-odds that the marker expression levels predict the outcome (control on the left vs. pregnancy on the right). Summary data are depicted, showing the 95% confidence interval. Markers in which the bar does not cross zero are significantly predictive of one state vs. the other. (D) Represents the linear discriminant analysis (LDA) for CD56dim NK cells between non-pregnant, pregnant, and post-partum samples from discovery cohort. (E,F) Markers predictive of CD56bright NK cells in control vs. pregnant women in discovery cohort were assessed by GLM with bootstrap resampling. The markers are listed on the y-axis and the x-axis represents the log-odds that the marker expression levels predict the outcome (control on the left vs. pregnancy on the right). Summary data are depicted, showing the 95% confidence interval. Markers in which the bar does not cross zero are significantly predictive of one state vs. the other. (G) Represents the linear discriminant analysis (LDA) for CD56bright NK cells between non-pregnant, pregnant, and post-partum samples from discovery cohort.