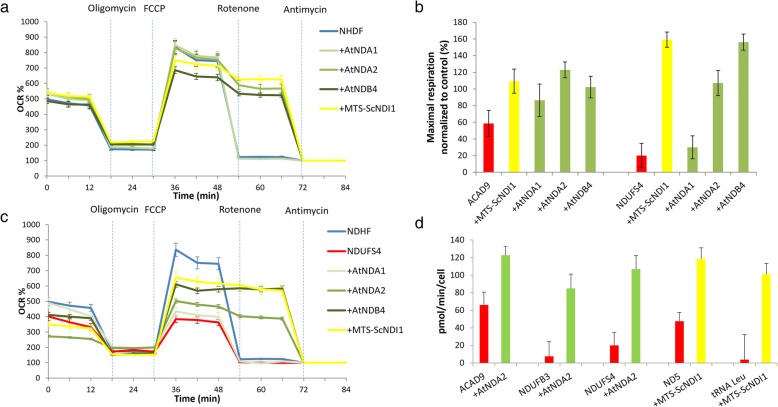
Fig. 2.
Oxygen consumption analysis: Oxygen consumption was evaluated using the Seahorse XF Analyzer; a Oxygen consumption rate (OCR) expressed as percent (%) of rate measurement 13 in control cells (NDHF) and in control cells transduced with alternative dehydrogenases from A. thaliana (+AtNDA1, +AtNDA2, +AtNDB4) and yeast (+MTS-ScNDI1); b OCR expressed as % of rate measurement 13 in NDHF, in NDUFS4-deficient cells (NDUFS4) and in patient cells transduced with alternative dehydrogenases from A. thaliana (+AtNDA1,+AtNDA2, +AtNDB4) and yeast (+MTS-ScNDI1); c Maximal respiration rate in CI deficient cells (carrying pathogenic variants in ACAD9 and NDUFS4), before and after transduction with AtNDA1, AtNDA2, AtNDB4 and MTS-ScNDI1. Values were normalized to maximal respiration of untransduced control cells; d Oxygen consumption rate (OCR) expressed as pmol O2/min/cell in cell lines presenting with CI-defect due to mutations in ACAD9, NDUFB3, NDUFS4, ND5, tRNALeu before and after transduction with alternative dehydrogenases from A. thaliana and yeast. Each cell line was measured at least twice in independent experiments. During experiment, four technical replicates were run for each cell line. Values are expressed as mean ± SD