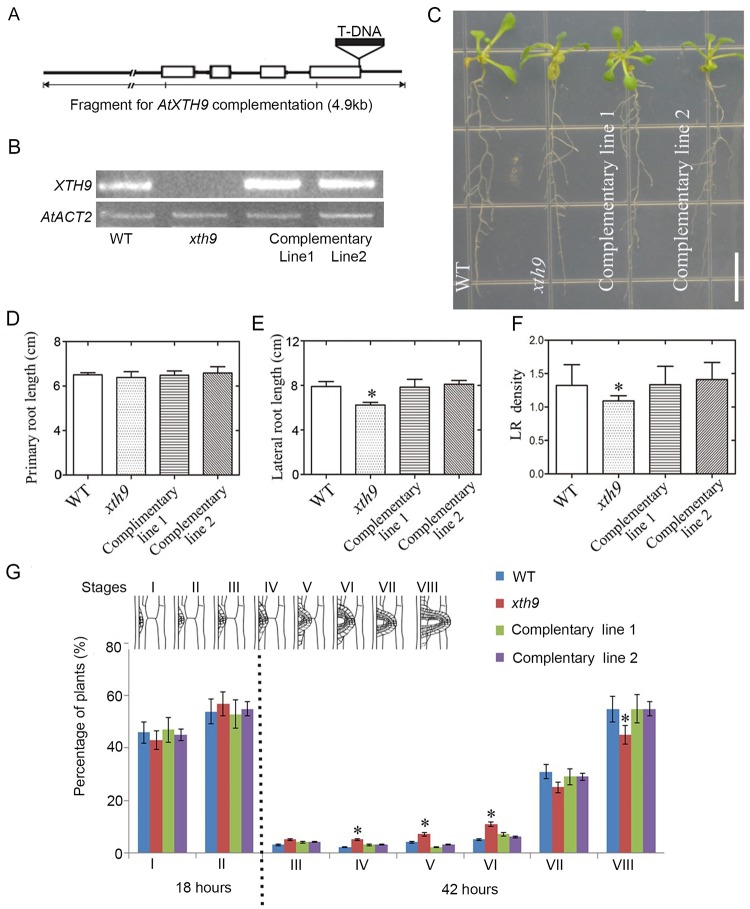
Fig 2. XTH9 gene mutation affects lateral root (LR) development.
(A) The genomic organization of the XTH9 gene. The white boxes show the positions and sizes of the XTH9 gene exons. The black box indicates the structure of the T-DNA, the site of which is indicated by the triangle. The genomic sequences used to complement the xth9 mutation are underlined with a thick line. (B) XTH9 transcript levels in wild-type (WT) (Col-0) plants, the xth9 mutant, and xth9 complementary lines. The mRNA abundance of the XTH9 gene in the roots was measured using RT-PCR in various genotype backgrounds of Arabidopsis. (C) Root phenotypes of WT plants, the xth9 mutant, and complementary lines. The plants were grown vertically on media for 10 days. The white bar indicates 1 cm. (D) Primary root length and (E) total LR length. (F) LR density analysis of WT plants, the xth9 mutant, and xth9 complementary lines. The error bars denote SDs (n = 16–21). (G) Phenotypic analysis of lateral root emergence (LRE) was achieved by synchronizing LR formation with a gravity stimulus for 18 and 42 hours. Compared with the WT plants, the xth9 mutants showed delayed LRE. Mutant plants transformed with a 4.9-kb-long full-length XTH9 genomic fragment exhibited a WT LRE phenotype for LR induction. The data are shown as percentages, and the error bars represent SDs (n = 15–17). At least 80–100 total LR primordia were observed for each plant, and the asterisks (*) indicate statistically significant differences (p<0.05).