Figure 2.
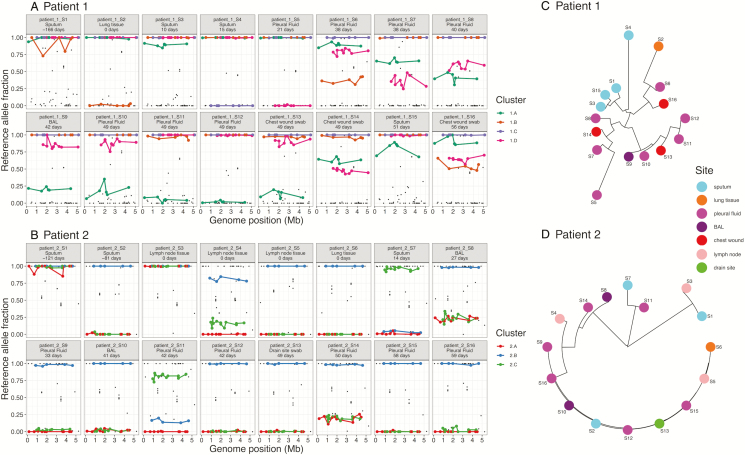
Linkage patterns of SNVs across samples suggest the presence of closely related subpopulations within patients. A and B, SNVs were grouped into clusters (colors) using an unsupervised clustering technique, showing clear patterns of abundance across samples (see Methods). The genome position was inferred by ordering de novo contigs against the Mycobacterium abscessus ATCC 19977 reference genome. C and D, The midpoint-rooted neighbor-joining trees are based on Euclidean distances between samples, using these clustered SNVs to show this variation within patients over time (numbers) and body sites (colors). Abbreviations: BAL, bronchoalveolar lavage; SNV, single-nucleotide variants.