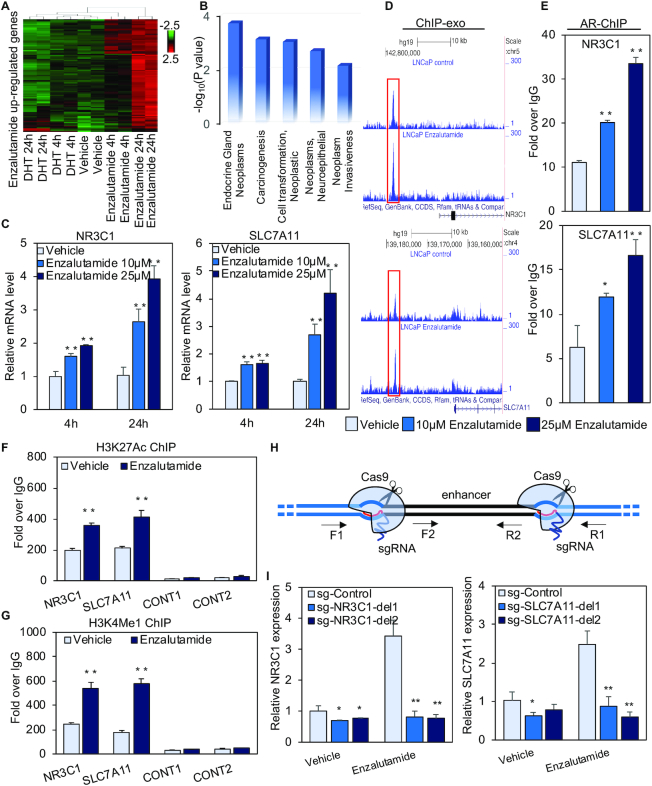
Figure 1.
Enzalutamide regulates global, cancer-relevant transcription. (A) A heatmap of enzalutamide upregulated genes. The gene expression (reads per kb per million mapped reads (RPKM)) values for each gene were normalized to the standard normal distribution to generate Z-scores. The scale bar is shown with the minimum expression value for each gene in green and the maximum value in red. (B) Genomatix Pathway containing enzalutamide upregulated genes. (C) Expression of NR3C1 and SLC7A11 was determined by quantitative RT-PCR after LNCaP cells were treated with vehicle or 10 μM or 25 μM enzalutamide for 4 or 24 h. (D) IGV shows AR ChIP-exo data at the NR3C1 and SLC7A11 loci. (E) AR ChIP validation of the NR3C1 and SLC7A11 loci in LNCaP cells treated with vehicle or 10 μM or 25 μM enzalutamide for 4 h. (F and G) The enrichment of H3K4Me1 (F) and H3K27Ac (G) within the NR3C1 and SLC7A11 loci in LNCaP cells treated with vehicle or with 25 μM enzalutamide for 4 h was determined by ChIP assays. (H) Schematic of CRISPR/Cas9-mediated deletion of the enhancers. The annealing sites for the primers used to validate deletion are indicated. (I) Expression of NR3C1 and SLC7A11 was measured by quantitative RT-PCR after cells were treated with vehicle or 25 μM enzalutamide for 24 h. sg-Control, a pair of sgRNAs that are predicted to not recognize any genomic regions; sg-del1 and sg-del2, two separate pairs of sgRNAs recognizing the boundaries of the enhancer region. RNA-seq was performed with two biological replicates (r > 0.99, Spearman's correlation). Statistical significance was assessed using the Student's t-test, *P < 0.05, **P < 0.01. Results were reported as the mean of two to four replicates, with error bars representing the standard deviation.