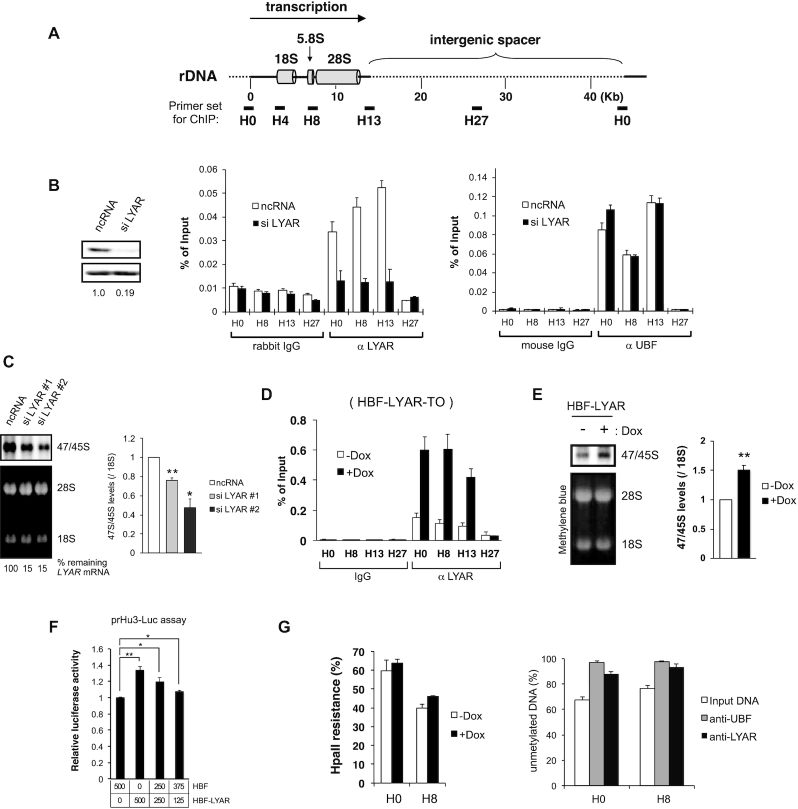
Figure 1.
LYAR is involved in rDNA transcription. (A) Schematic diagram of a single human rDNA genic region (rDNA repeats are separated by an intergenic spacer). Primer sets (short bars: subregions H0, H8, H13, and H27) used for ChIP analysis are indicated along with their approximate positions relative to the transcription start site (0 kb). (B) ChIP analysis of UBF and LYAR binding to rDNA loci in HeLa cells transfected with scRNA (control) or siRNA specific for LYAR. Nonspecific mouse IgG or rabbit IgG was used as the antibody control. The chromatin-immunoprecipitated (ChIPed) DNA was quantified by qPCR using the primer sets indicated in A. The graph shows the amount of ChIPed DNA (% of input). Data represent the mean ± SEM of three independent experiments. The efficiency of LYAR knockdown was assessed with immunoblotting. (C) Metabolic labeling (4-thiouridine) of newly synthesized 47/45S pre-rRNA in HeLa cells upon LYAR knockdown. The RNA extracted for the cells treated with siRNA #1, siRNA #2 or ncRNA specific for LYAR (or ncRNA, control), was biotinylated and then subjected to agarose gel electrophoresis under denaturing condition and northern blotting. Signals for 47/45S pre-rRNA were detected by chemiluminescence. 28S and 18S rRNAs were used as loading controls (stained with methylene blue). The graph shows the relative band intensities of biotin-labeled 47/45S pre-rRNA normalized to that of 18S rRNA. Data reflect the mean ± SEM of three independent experiments. *P < 0.05 (paired t-test). LYAR mRNA levels, normalized to GAPDH mRNA, were assessed by reverse transcription-qPCR. (D) ChIP analysis of LYAR binding to rDNA loci (H0, H8, H13, H27) in HBF-LYAR-TO cells with or without HBF-LYAR induction via Dox. Nonspecific rabbit IgG was used as the antibody control. The graph shows the amount of ChIPed DNA (% of input). Data reflect the mean ± SEM of three independent experiments. (E) Metabolic labeling (4-thiouridine) of newly synthesized 47/45S pre-rRNA in HBF-LYAR-TO cells with or without Dox treatment. The pre-rRNA was biotinylated and then subjected to agarose gel electrophoresis under denaturing condition and northern blotting. Signals for 47/45S pre-rRNA were detected by chemiluminescence. 18S rRNAs were used as loading controls (stained with methylene blue). The graph shows the relative band intensities of 2 biotin-labeled 47/45S pre-rRNA normalized to that of 18S rRNA. Data reflect the mean ± SEM of six independent experiments. **P < 0.01 (paired t-test). (F) prHu3-Luc assay showing the effect of LYAR on RNAP I–dependent transcription. 293T cells were co-transfected with the prHu3-Luc reporter gene (firefly luciferase) and internal control vector pRL-TK (Renilla luciferase) along with the indicated amount of HBF and/or HBF-LYAR expression vectors, and luciferase activities were measured. The graph shows the ratio of the luciferase activities (firefly/Renilla) along with the corresponding HBF-LYAR expression levels or levels of HBF only. Data represent the mean ± SEM of three independent experiments. *P < 0.05, **P < 0.01 (paired t-test). (G) Methylation of rDNA in HBF-LYAR-TO cells was assessed with the HpaII resistance assay. HBF-LYAR-TO cells were treated with Dox for 24 h and then subjected to HpaII resistance assay. ChIP-CHOP experiment, in which the rDNA promoter-proximal DNA that had been subjected to ChIP with anti-LYAR or anti-UBF was subjected to HpaII digestion to determine whether LYAR associates with transcriptionally active, unmethylated rDNA repeats.