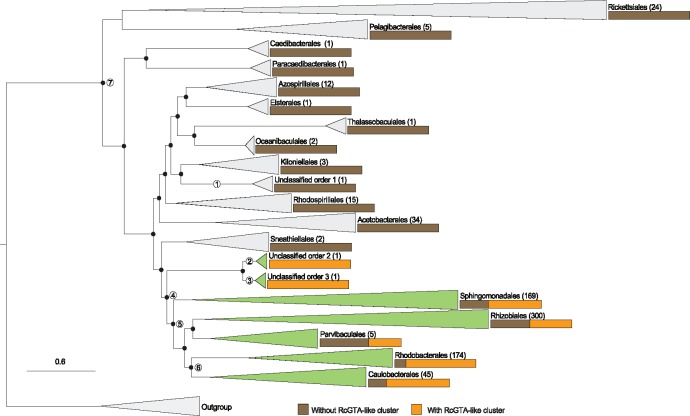
Fig. 3.
—Distribution of the detected RcGTA-like clusters across the class Alphaproteobacteria. The presence of RcGTA-like clusters is mapped to a reference phylogenetic tree that was reconstructed from a concatenated alignment of 83 marker genes (See Materials and Methods and supplementary table S9, Supplementary Material online). The branches of the reference tree are collapsed at the taxonomic rank of “order,” and the number of OTUs within the collapsed clade is shown in parentheses next to the order name. Orange and brown bars depict the proportion of OTUs with and without the predicted RcGTA-like clusters, respectively. The orders that contain at least one OTU with an RcGTA-like cluster are colored in green. Nodes 1–3 mark the last common ancestors of the unclassified orders. Node 4 marks the lineage where, based on this study, the RcGTA-like element should have already been present. Nodes 5 and 7 mark the lineages that were previously inferred to represent last common ancestor of the RcGTA-like element by Shakya et al. (2017) and Lang and Beatty (2007), respectively. Node 6 marks the clade where RcGTA-like elements are the most abundant. The tree is rooted using homologs from Escherichia coli str. K12 substr. DH10B and Pseudomonas aeruginosa PAO1 genomes. Branches with ultrafast bootstrap values >=95% are marked with black circles. The scale bar shows the number of substitutions per site. The full reference tree is provided in the FigShare repository.