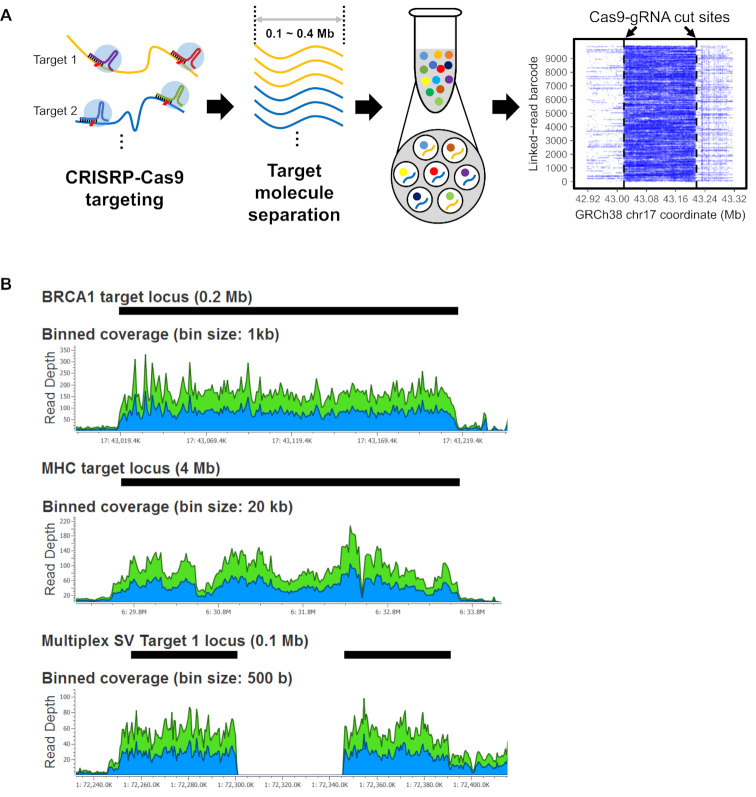
Figure 1.
CATCH targeting and linked read sequencing of HMW DNA. (A) Overview of the process is illustrated. First, guide RNAs target and cut multiple genomic regions of interest. Second, target HMW DNA within the specific size range is isolated by an electrophoresis-based process. At last, the target DNA is used for linked read library preparation and sequencing. The alignment of barcode linked reads shows how sequence coverage is increased across the target segment. In the alignment plot, the X-axis indicates the reference coordinates and the Y-axis shows different barcodes representing individual HMW molecules. Dashed vertical lines indicate Cas9-gRNA cut sites. (B) Sequencing coverage for the target regions is shown for the three assays. For Assays 1 and 2, BRCA1-R2 and MHC-30 libraries are shown. For Assay 3, an example of a homozygous deletion (SV1) is shown. Black bars indicate the target regions. Blue and green areas in plots indicate coverage for forward and reverse reads, respectively.