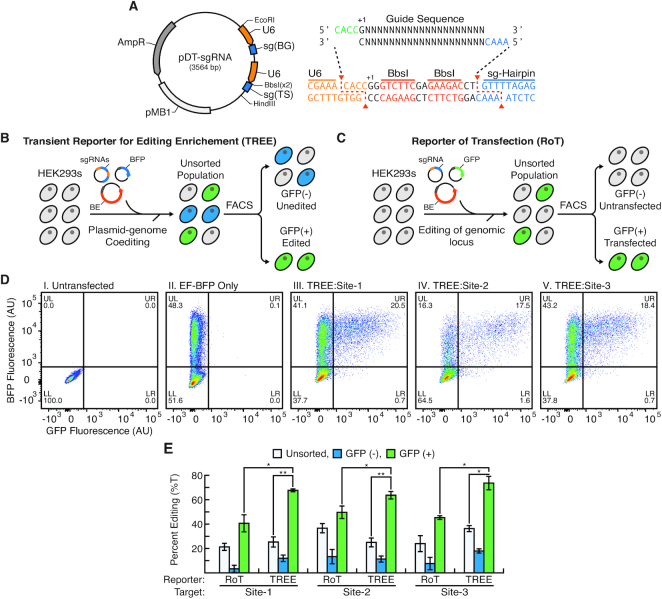
Figure 3.
Enrichment of base-edited cell populations using TREE. (A) Plasmid map of pDT-sgRNA vector that contains sg(BG) and sg(TS). Expression for both sgRNA cassettes is driven by separate U6 promoters (orange arrows). The BbsI restriction sites allow for direct restriction enzyme-based cloning of new target sites. (B) Schematic for enrichment of edited cells using TREE. HEK293 cells are co-transfected with pEF-BFP, pCMV-BE4-Gam and pDT-sgRNA vectors. After 48 h post-transfection, flow cytometry is used to sort cell populations into GFP-positive and -negative fractions. (C) Schematic for enrichment of edited cells using reporter of transfection (RoT). HEK293 cells are co-transfected with pEF-GFP, pCMV-BE4-Gam and sg(TS) vectors. After 48 h post-transfection, flow cytometry is used to sort cell populations into GFP-positive and -negative fractions. (D) Representative flow cytometry plots of (i) untransfected HEK293 cells and (ii) HEK293 cells transfected with pEF-BFP only as well as HEK293 cells in which TREE was applied targeting (iii) Site-1, (iv) Site-2 and (v) Site-3. (E) Quantification of base editing efficiency at Site-1, Site-2 and Site-3 in GFP-positive, GFP-negative and unsorted cell populations isolated using TREE- or RoT-based enrichment strategies. n = 3; * = P < 0.05, ** = P < 0.01.