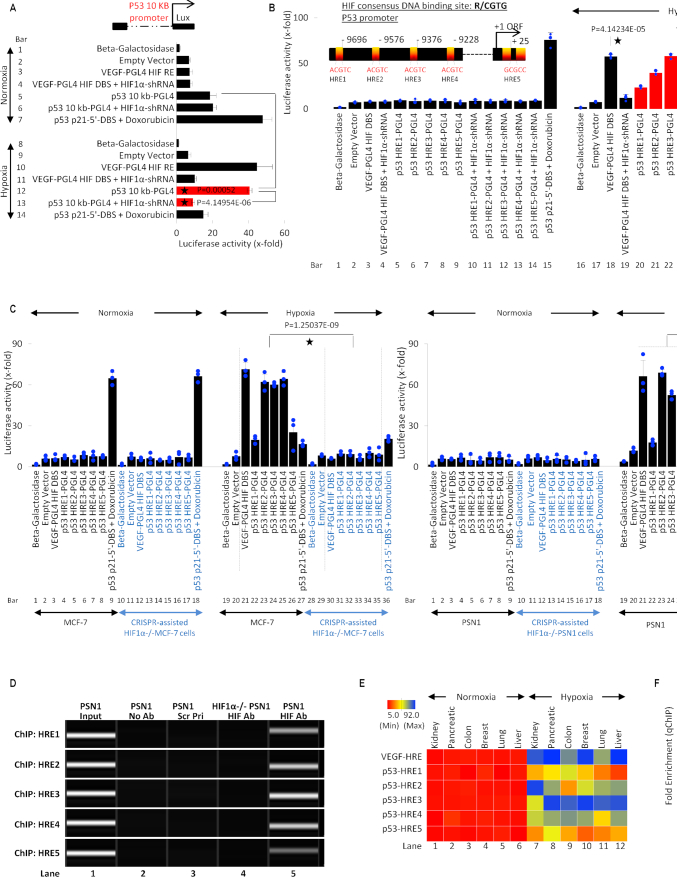
Figure 4.
HIF-1α transcriptionally regulates p53. (A) Luciferase assay was used to determine hypoxia-induced and HIF-1α-dependent activation of full-length p53 (10kb) promoter, a known HRE in VEGF promoter was cloned in PGL4 promoter and used as a positive control. Normoxia and hypoxia p53 WT MCF-7 cells were transfected with VEGF-PGL4-HRE or p53 10kb-PGL4 alone or co-transfected with HIF-1α-shRNA under and harvested for luciferase assay. A known p53 DBS in p21 promoter, p53 p21–5′-DBS-pGL4 was used as a positive control in doxorubicin-treated normoxic MCF-7 cells. Empty vector or Beta-galactosidase (β-Gal) served as normalization controls; error bars represent S.D., n = 3. Two-sided Student's t-test was used for analysis; all P values are labeled on the figure. (B) Luciferase assay was used to assess the activity of 5 putative HREs in the p53 promoter, as depicted in the model. p53 WT MCF-7 normoxic and hypoxic cells were transfected with VEGF-PGL4 vector, as a positive control for HIF-1α activity, or each p53-HRE-PGL4 vectors with or without HIF-1α-shRNA. Doxo-treated MCF-7 positive cells transfected with p53 p21-5′-DBS-pGL4 vector serves as positive control under normoxic conditions. The five p53-HREs sites display hypoxia-inducible luciferase activity that is blocked by shRNA-mediated HIF-1α knockdown. Empty vector or Beta-galactosidase (β-Gal) served as normalization controls; error bars represent S.D., n = 3. Two-sided Student's t-test was used for analysis; all P values are labeled on the figure. (C) Luciferase assay was used to assess the 5 HREs in p53 promoter under normoxia and hypoxia in HIF-1α-WT/KO MCF-7 and PSN1 cells. All cells were treated as described in (B) and harvested for luciferase assay. In both HIF1-α-WT MCF-7 (WT p53) and PSN1 (MT p53) cell lines, hypoxia leads to robust p53 promoter activity at each HRE (lanes 22–26, left and right panel) compared to normoxia. Hypoxic induction of HRE promoter activity is lost in HIF-1α−/− MCF-7 and PSN1 cells (lanes 31–35, left and right panel), indicating hypoxic induction of p53 promoter activity is HIF-1α-dependent. Empty vector or Beta-galactosidase (β-Gal) served as normalization controls; error bars represent S.D., n = 3. Two-sided Student's t-test was used for analysis; all P values are labeled on the figure. (D) The ChIP-PCR assay was used to determine if HIF-1α binds to the five predicted HREs in p53 promoter in hypoxic WT PSN1 and HIF-1α−/− PSN1 cells. Lane 1: chromatin input. Lane 2: No Antibody negative control. Lane 3: a scrambled primer (Scr Pri) as a control for non-specific DNA PCR amplification. Lane 4: HIF-1α−/− cells as a negative control for HIF-1α protein binding. Lane 5: binding of HIF-1α protein to each of the 5 HREs is observed, n = 3. (E) Heat map depicts HIF-1α binding to the 5 HREs in p53 promoter measured by ChIP-qPCR in normoxic and hypoxic regions of kidney, pancreatic, colon, breast, lung and liver patient tumors. ChIP for HIF-1α binding to the VEGF-HRE was included as a positive control to confirm HIF-1α activation and binding activity. Variable HIF-1α binding to p53-HREs was detected in hypoxic regions, but not normoxic regions, from all cancer types. In this figure, each block represents the values from a gradient scale of low (red), medium (yellow), and high (blue) level of expression, n = 3 (biological replicates). (F) Box plot summary of ChIP-qPCR enrichment for each HRE in cancer hypoxic regions as described in (E). Fold enrichment depicts HIF-1α binding to VEGF-HRE or HRE 1–5 in p53 promoter, in the hypoxic regions relative to HIF-1α binding in the normoxic regions, error bars represent S.D., n = 3.