Figure 2.
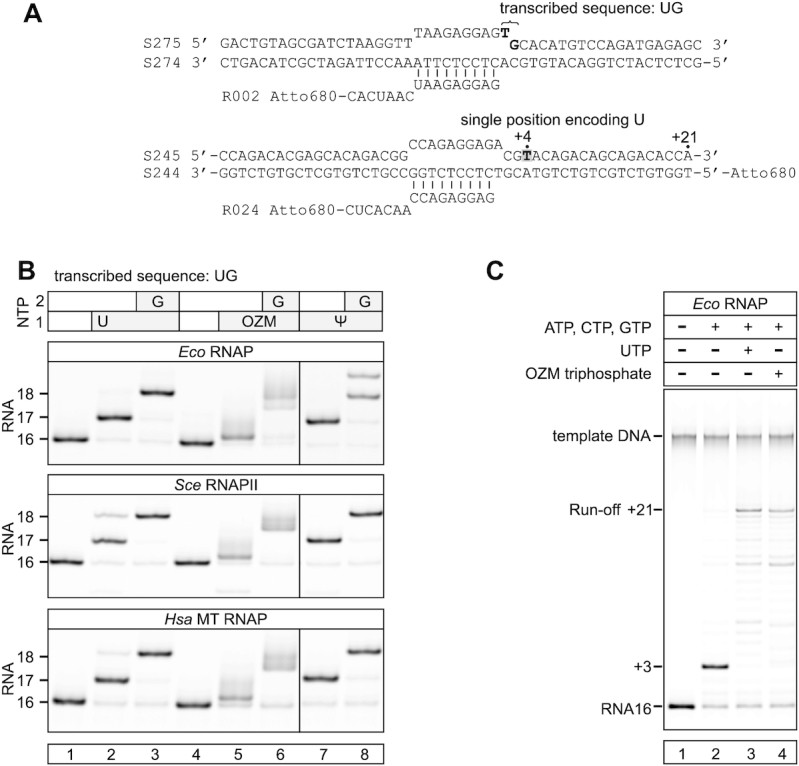
Eco RNAP, Sce RNAPII and Hsa MT RNAP efficiently incorporate OZM in place of uridine. (A) The nucleic acid scaffolds employed in the experiments in (B) and (C). (B) Incorporation of OZM in place of uridine: the initial TECs (lanes 1 and 4; top schematics in (A)) were supplemented with UTP and GTP (lanes 2 and 3), OZM triphosphate and GTP (lanes 5 and 6), ΨTP and GTP (lanes 7 and 8). NTPs were added at 20 μM and the reactions were incubated for 2 min at 25°C. RNAs were resolved on 25% urea PAGE. Fractional misincorporations (additional bands) are evident in several lanes. (C) OZM triphosphate allows efficient transcription of the sequence position encoding uridine: the initial TEC (lane 1, bottom schematics in (A)) was supplemented with ATP, CTP and GTP (lane 2) and additionally with UTP (lane 3) or OZM triphosphate (lane 4). NTPs were added at 100 μM and the reactions were incubated for 15 s at 25°C. The limited read through the position encoding uridine in the absence of UTP and OZM triphosphate (lane 2) was likely due to the misincorporation of CMP in place of UMP. Each assay was performed in triplicate. Pixel counts were linearly scaled to span the full 8-bit grayscale range within each gel panel.
