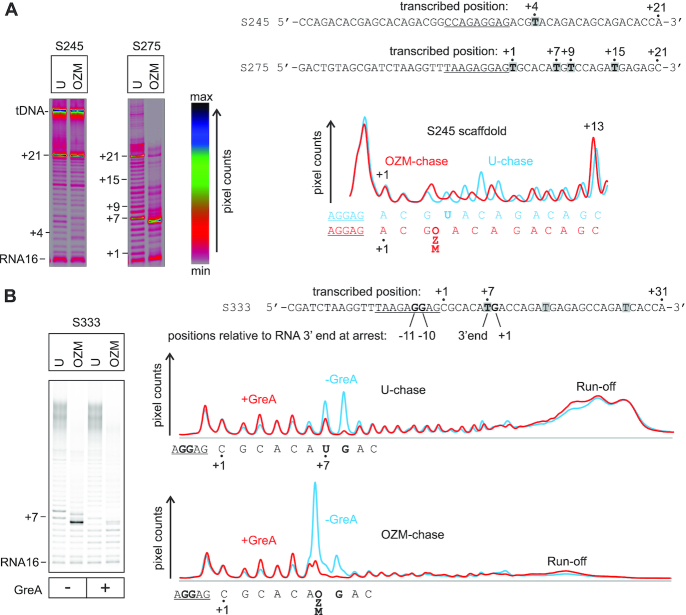
Figure 6.
Transcription through a template encoding an OZM-responsive arrest site by Eco RNAP. TECs were assembled using the scaffolds shown to the right of the gel panels (only the non-template DNA strands are shown) and chased with 100 μM ATP, CTP, GTP and UTP or OZM-triphosphate for 5 min at 25°C in the presence or absence of 2 μM GreA. The sequence corresponding to the annealing region of the RNA primer is underlined, thymidines in the transcribed region are highlighted. Each assay was performed in triplicate. (A) Transcription through two representative short templates. The lane profiles depict the effect of OZM on transcription of the S245 template (left gel panel). Pixel counts were linearly scaled to span 256 gradations within each gel panel. Gels were pseudocolored using the RGB lookup table shown to the right of the gel panels to visualize the low intensity bands. (B) Transcription through the simplified and longer derivative of the S275 template (right gel in A) that encodes the OZM-responsive arrest site. The OZM-responsive arrest site at +7 encodes the main determinants of a consensus pause: pyrimidine at the RNA 3′ end, purine at +1 and Gs at −11 and −10, these determinants are shown in bold. Pixel counts were linearly scaled to span the full 8-bit grayscale range. Lane profiles quantified from the gel are presented on the right. The RNA rescued from the arrest site by GreA in OZM-chase does not quantitatively reappear upstream or downstream of the arrest site (see the text for details and possible explanations).