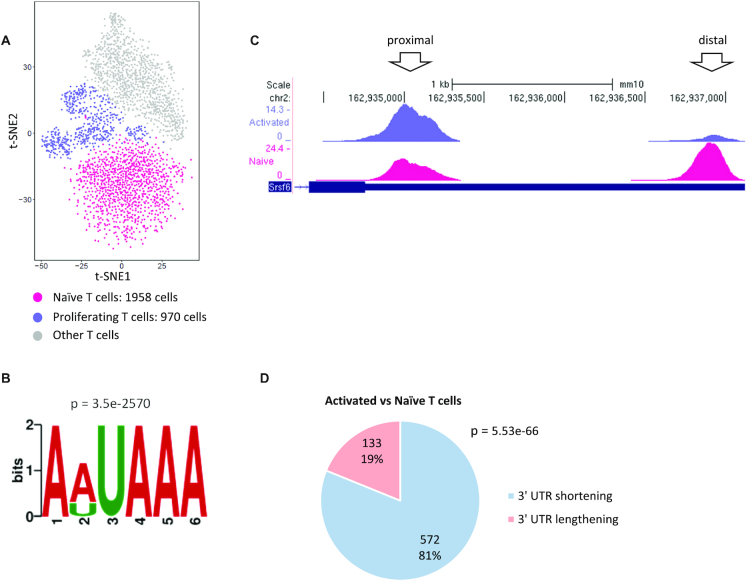
Figure 2.
Analysis of APA modulation in activated T cells. (A) t-SNE plot of all the T cells from wild type mice identified by Pace et al. (44). We analysed the clusters of proliferating T cells (970 cells; purple) and naïve T cells (1,978 cells; pink), defined in the original publication. (B) The top-scoring signal detected by de novo motif analysis of the 9611 3′ UTR peaks corresponds to the canonical PAS motif and its main variant. (Sequences used for this analysis spanned the region from 30 nt upstream to 120 nt downstream of the peaks 3′ edge.) (C) An example of a gene that shows significant 3′ UTR shortening in activated T cells (top track) compared with naïve T cells (bottom track). (D) A pie chart for the distribution of 3′ UTRs with exactly two peaks that showed a significant change in pA site usage in the comparison between proliferating and naïve T cells. (P-value calculated using single-tailed binomial test.)