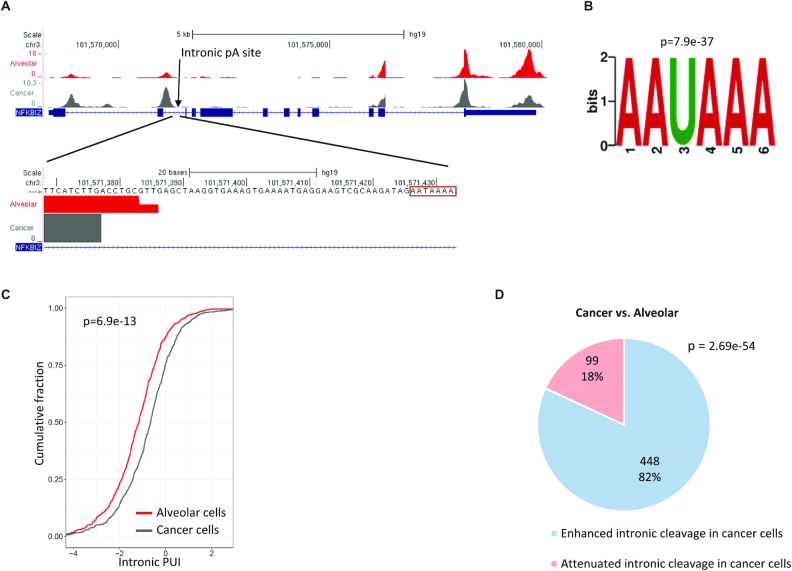
Figure 6.
Analysis of intronic APA in lung tumour sample. (A) An example of a gene that shows, in the comparison between cancer and normal cells, a significant increase in the usage of an intronic pA site. This intronic site harbours the canonical AAUAAA signal at the expected location. (Note that in addition to the enhanced cleavage at the intronic pA site, this gene also shows the typical increased usage of the 3′ UTR proximal pA site in cancer cells compared with the normal ones.) (B) The PAS signal was the top-scoring motif detected by de novo motif analysis applied to the 3057 intronic peaks that were left in the analysis after the removal of putative internal priming peaks. (C) Cumulative distribution of the Intronic PUI index in alveolar cells (red), and cancer cells (dark grey) (P-value calculated using single-tailed Wilcoxon test). The shift to the right of the cancer cells’ curve reflects enhanced cleavage at intronic pA sites. (D) A pie chart illustrating the distribution of intronic pA sites that showed a significant change in their usage in the comparison between cancer and alveolar cells. In blue and pink, respectively, are the proportions of intronic pA sites with enhanced or attenuated usage in cancer cells (P-value calculated using single-tailed binomial test).