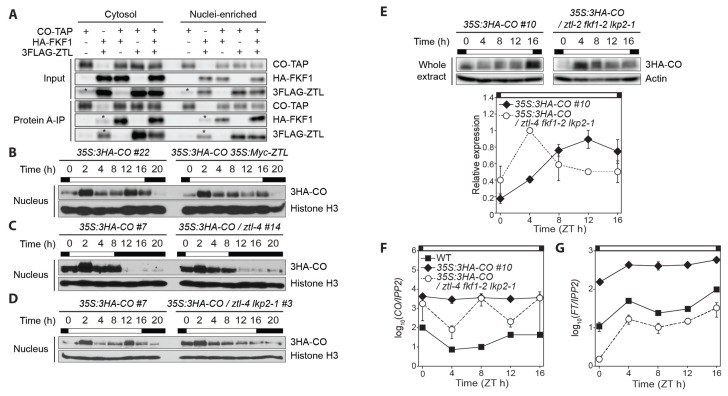
Fig. 3. GI and the ZTL group members regulate the daytime CO protein stability.
(A) Interactions among CO, FKF1, and ZTL in the cytosolic and nuclei-enriched fractions. N. benthamiana leaves constantly expressing CO-TAP, HA-FKF1, and 3FLAG-ZTL proteins were collected 3 days after infiltration. Asterisks denote non-specific bands. (B–E) Immunoblot assays for diel expression profiles of 3HA-CO protein in Arabidopsis transgenic plants. Similar expression patterns were observed in two biological replicates except (E). Nuclear 3HA-CO protein signals were detected by anti-HA antibody, and histone H3 protein was used for an internal control in the nucleus. (B) Long day-grown 35S:3HA-CO #22 and 35S:3HA-CO 35S:Myc-ZTL plants were harvested on day 10 for nuclei isolation. (C) The 35S:3HA-CO #7 and 35:3HA-CO / ztl-4 #14 transgenic plants were grown in short days for 14 days. (D) The 35S:3HA-CO #7 and 35S:3HA-CO / ztl-4 lkp2-1 #3 plants were collected at the time points indicated in long days. (E) The daytime expression of 3HA-CO protein in long days was compared between two CO overexpression lines in wild type and the ztl-4 fkf1-2 lkp2-1 mutant background. Immunoblot images (the upper panel) and quantified 3HA-CO protein abundance (the lower panel) in whole protein extract are shown. Actin protein was used as a loading control for normalization. A direct comparison between two CO transgenic plants is not valid. Means ± SEM were calculated from three independent replicates. (F and G) CO (F) and FT (G) mRNA levels in 10-day-old wild type, 35S:3HA-CO #10, and 35S:3HA-CO / ztl-4 #14 seedlings grown in long days. The mRNA levels were measured by quantitative real-time polymerase chain reaction and normalized against IPP2 gene. Daytime gene expression profiles are represented in a logarithmic scale. Means of three biological trials ± SEM.