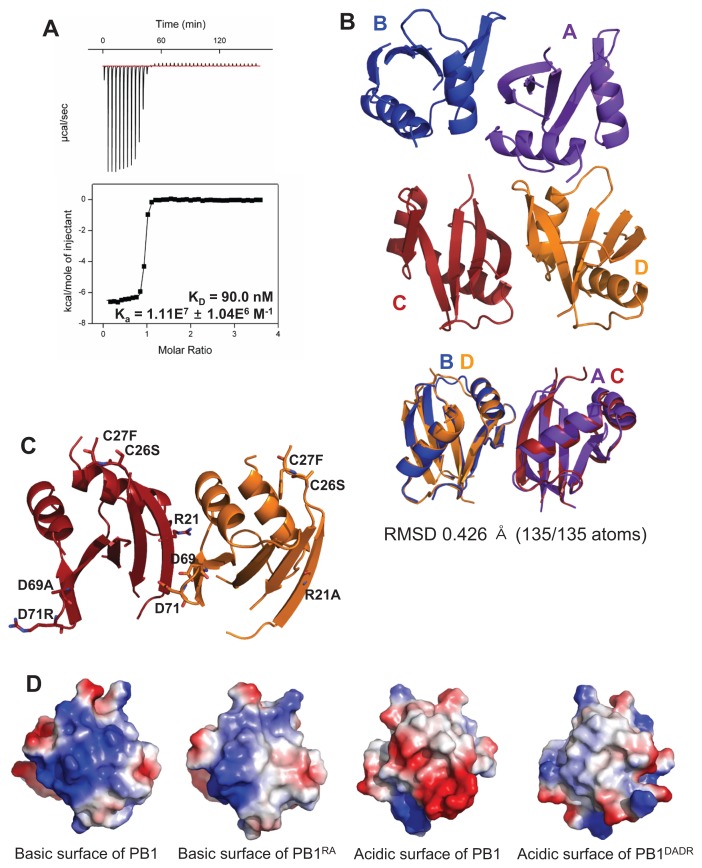
Fig. 1. Structural analysis of the crystal structure of the PB1 homo-dimer.
(A) The binding affinity between PB1RA and PB1DADR was measured by ITC. (B) The overall crystal structure of the PB1 domain has four chains as two dimers in the asymmetric unit. These two dimers are superposed. (C) The mutated residues are presented as stick and these mutations were located on surface of PB1 homo-dimer. (D) The basic and acidic surfaces of C and D chains are presented by vacuum electrostatics models. The mutated surfaces have less color than the wild type.