Fig. 1. Generation and characterization of myeloid-specific Trib1 mouse strains.
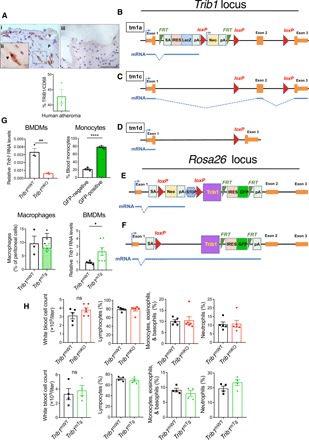
(A) Representative immunohistochemistry image of human atherosclerotic plaque (P). Red, TRIB1; brown, CD68+. (ii) Magnification (×40) of boxed area. Arrowhead highlights a double-positive cell. Quantification (mean ± SD) of three patient samples. (iii) Isotype control (scale bar, 50 μm). (B) Targeting construct used to produce the null, conditional-ready/floxed (tm1c) and conditional-null (tm1d) Trib1 alleles. Predicted transcripts below. FRT, flippase recognition target; SA, splice acceptor; pA, polyadenylation motif; IRES, internal ribosome entry site; LacZ, β-galactosidase; Neo, neomycin resistance gene. (C) Trib1mWT allele following removal of the “gene-trap” cassette. (D) Conditional-null Trib1 allele was produced by crossing tm1c and Cre-expressing mice. (E) Construct used to produce Trib1mWT and Trib1mTransgenic (Tg) mice. (F) Cre-mediated excision of the STOP cassette produces a bicistronic Trib1-eGFP transcript. Bent arrow, indicates transcription from endogenous Rosa26 promoter. (G) Trib1 RNA (relative to Actb) in BMDMs from homozygous tm1c (i.e., Trib1mWT) and Trib1mKO mice (n = 3 per group). eGFP expression in monocytes of three Trib1mTg mice and peritoneal macrophages from specified mice (n = 3 per group). Trib1 RNA levels (relative to Trib1mWT) in BMDMs of Trib1mTg (n = 5 to 7 per group). (H) Blood cell counts of mixed-gender Trib1mKO (top) and Trib1mTg (bottom) and their respective WT littermates (n = 5 to 6 per group). Data are means ± SEM. Significances were determined by Student’s t test, *P < 0.05, **P < 0.01, and ****P < 0.0001. ns, non-significant.
