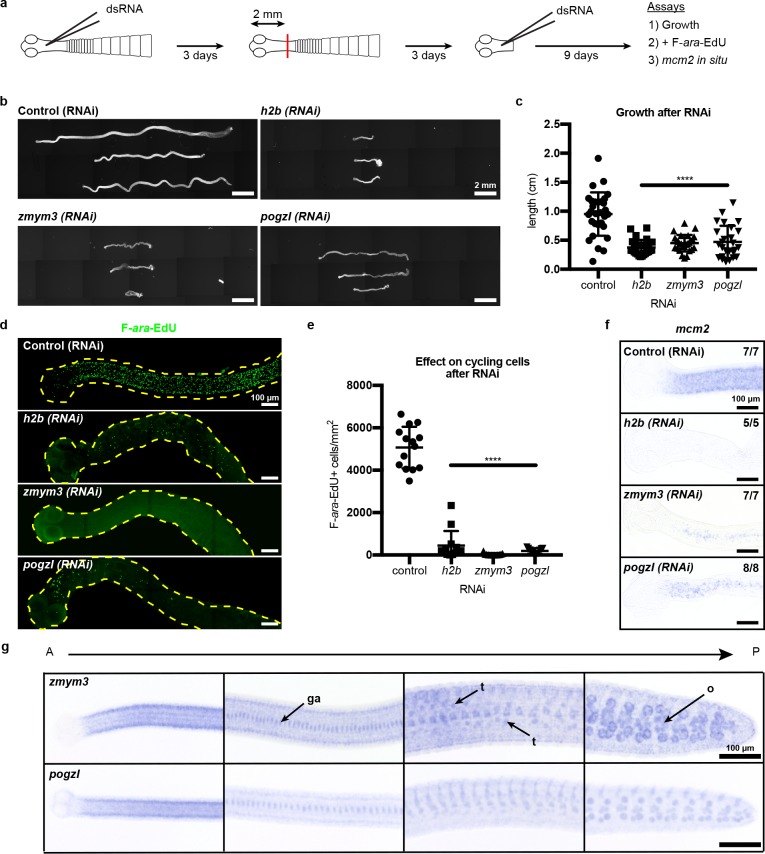
Figure 4. RNAi to identify genes required for growth and regeneration in H. diminuta.
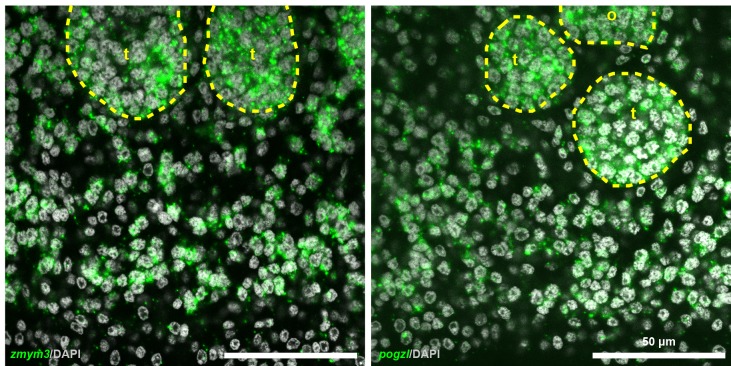
(a) Schematic of RNAi paradigm. (b) DAPI-stained worms after RNAi knockdown of h2b, zmym3, and pogzl. (c) Quantification of worm lengths after RNAi. Error bars = SD, N = 3–4, n = 26–37, one-way ANOVA with Dunnett’s multiple comparison test compared to control. (d-e) Maximum-intensity projections (d) and quantification (e) of cycling-cell inhibition after 1 hr F-ara-EdU uptake following RNAi. Worms with degenerated necks were excluded from analysis. Error bars = SD, N = 3, n = 11–14, one-way ANOVA with Dunnett’s multiple comparison test compared to control. (f) mcm2 WISH on worm anteriors after RNAi. (g) WISH of zmym3 and pogzl sampled from anterior to posterior of adult 6-day-old worms. ga: genital anlagen; t: testis; o: ovary.
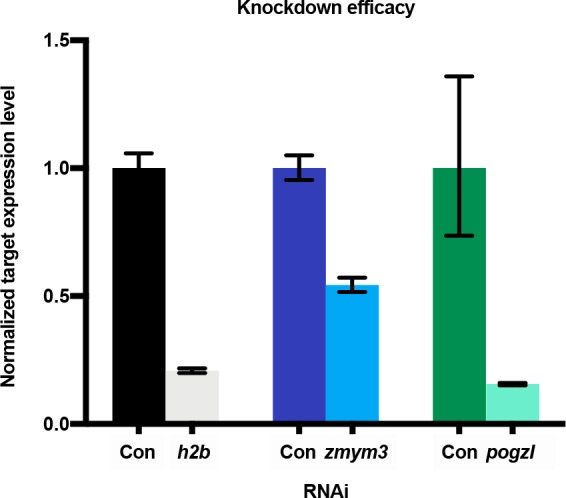
Figure 4—figure supplement 1. Validation of target gene knockdown by quantitative PCR.