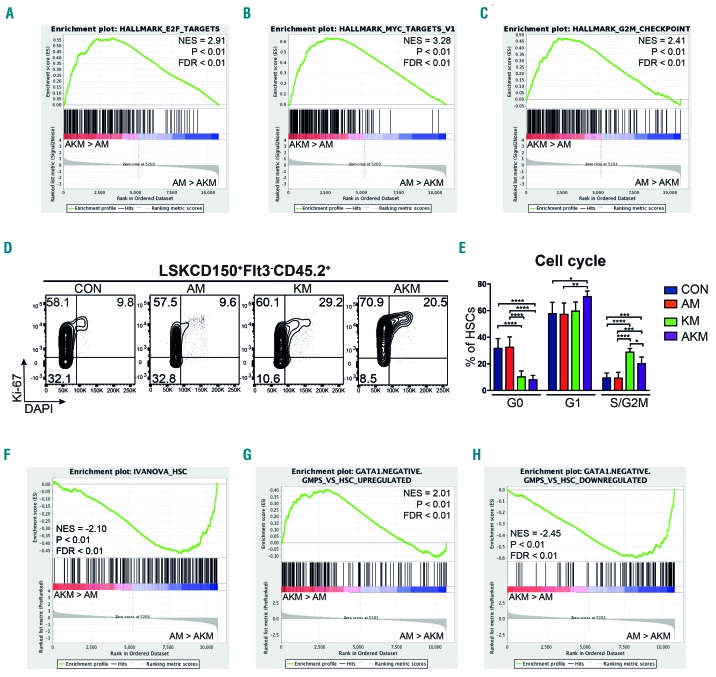
Figure 4.
Hematopoietic stem cells (HSC) co-expressing Aml1-ETO and K-RasG12D are characterized by loss of quiescence and HSC-associated gene expression. (A-C) Bulk CD45.2 LSKCD150+Flt3− cells were subjected to RNA sequencing (5-6 biological replicates per genotype in two independent experiments). Gene set enrichment analysis (GSEA) of AKM versus AM HSC for E2F targets (A), Myc targets (B), and genes associated with G2M checkpoint (C). (D) Representative FACS plots showing cell cycle analysis of CD45.2 LSKCD150+Flt3− phenotypic HSC from the bone marrow (BM) of recipients of CON (n=6 recipient mice in 2 independent experiments), AM (n=9 recipient mice in 3 independent experiments), KM (n=4 recipient mice in 2 independent experiments), and AKM FL (n=6 recipient mice in 3 independent experiments). (E) Percentage of BM CD45.2 LSKCD150+Flt3− cells at each cell cycle stage. The results were analyzed using multiple comparison ANOVA. The results are presented as mean±Standard Deviation. *P<0.05; **P<0.01; ***P<0.001. (F-H) GSEA analysis of AKM versus AM HSC for HSC gene signature (F), genes up-regulated in granulocyte-monocyte progenitor (GMP) that lack Gata1 expression compared to HSC (G), and genes down-regulated in GMP that lack Gata1 expression compared to HSC (H). NES: normalized enrichment score; FDR: false discovery rate.