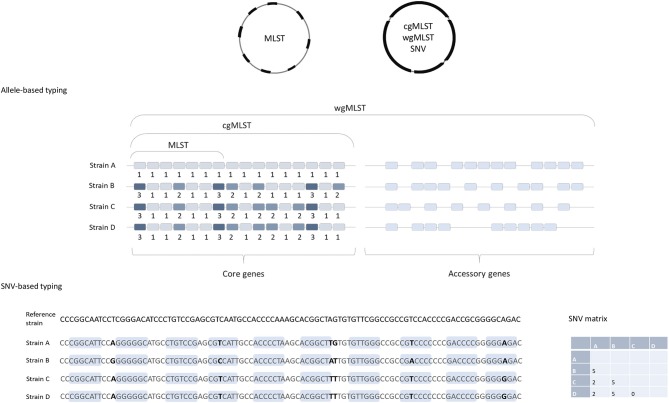
Figure 1.
Comparison of allele-based and SNV-based typing approaches. cgMLST, wgMLST, and SNV approaches are based on the genome-wide analysis and MLST includes only seven housekeeping genes. Note that strains B, C, and D are identical in MLST approach (same allelic profile in seven genes) and both are the same MLST-ST; but they would differ in cgMLST, strain C and D having identical cgMLST allelic profile, and strain B differs from C and D in three additional genes. In the SNP-based approach, short reads are aligned to a reference genome and the nucleotide differences in both coding (light blue boxes) and non-coding regions (excluding horizontally acquired elements and putative recombination regions) are determined. The number of SNV differences between the pairs of isolates is presented in the matrix on the right.