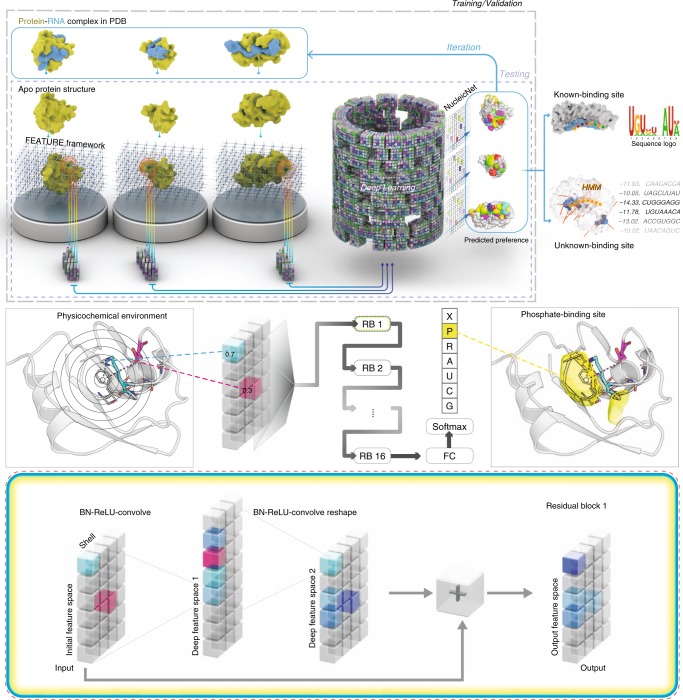
Fig. 1.
Overview of NucleicNet. Top panel: training strategy and utilities of NucleicNet. Ribonucleoprotein structures in the PDB are stripped of their bound RNA. Grid points are placed on the surfaces of the proteins. Each of these grid points (locations) is analyzed by the FEATURE program for their surrounding physiochemical environment, which is encoded as a feature vector. Binding locations for all six classes of RNA constituents and non RNA-binding sites are labeled accordingly. The labels together with their respective vectors are compiled; this formulates the training input for a deep residual network. Parameters in the network are iteratively updated by backpropagation of errors and are trained to differentiate the seven classes. Once training is complete, the learned model can then be used to predict binding sites of RNA constituents for any query protein structure surface. Downstream applications of the prediction outcome includes production of logo diagrams for RBPs and a scoring interface for any query RNA sequence. Middle panel: physicochemical environment accession and introduction on the residual network. In the FEATURE vector framework, physicochemical environment is perceived by accounting properties on atoms of a protein within 7.5 Å of a grid point in a radial distribution setup. As such, space surrounding each grid point is divided into six concentric shells of spheres and, for each of these shells, 80 physicochemical properties (e.g., negative/positive charges, partial charges, atom types, residue types, secondary structure of the possessing residue, hydrophobicity, solvent accessibility, etc.) were accounted resulting in a tensor of dimension 6×80. The tensor is then transformed by a deep residual network with 16 sequential residual blocks. After that, the final residual block is connected to a fully connected layer with a softmax operation to assess binding probability of each class on that location. Bottom panel: we illustrate the principle operations in a residual network, namely batch normalization (BN), rectified linear unit (ReLU), locally connected network, and the quintessential skip connection, which adds the initial input back to the penultimate output layer