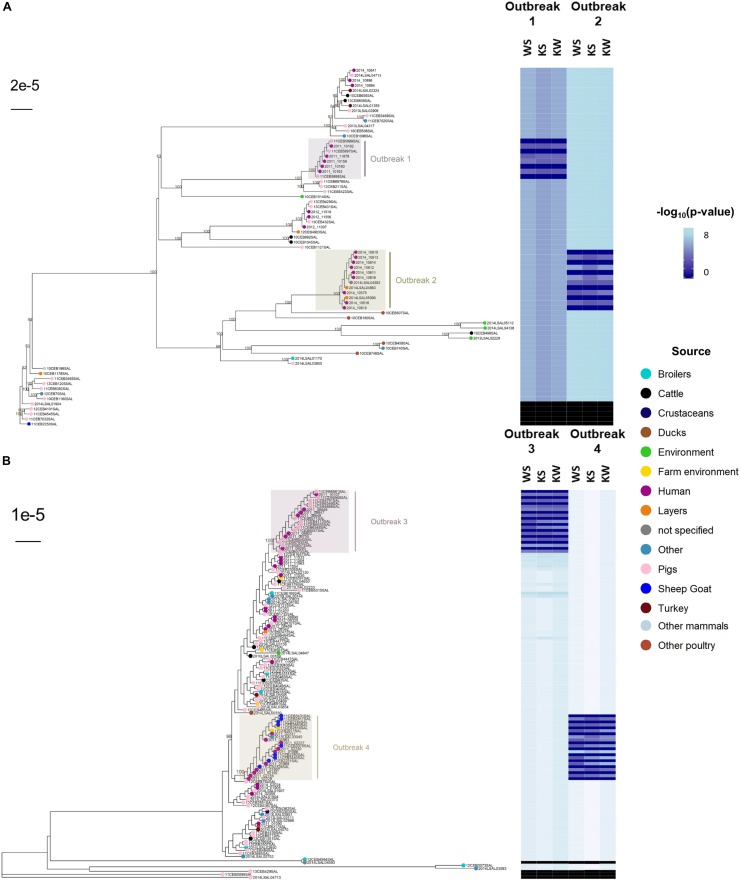
FIGURE 4.
Phylogenetic inference based on coregenome single nucleotide polymorphisms (SNPs) identified in 192 Salmonella enterica subsp. enterica during outbreaks in France caused by serovars Typhimurium (A: outbreaks #1 and #2; n = 66) and S. 1,4,[5],12:i:- (B: outbreaks #3 and #4; n = 126) and related p-values from non-parametric tests WS (i.e., differences of median values), KS (i.e., differences in distributions) and KW (i.e., differences of mean ranks) aiming to access statistical differences of pairwise SNP differences (i.e., approaches ‘SNPs-1’ including recombination events). The R script ‘matrix2association’ estimates statistical differences between two lists of pairwise differences existing across all genomes known to be involved in a studied outbreak (i.e., outbreak test set: TS) and between these genomes and a tested genome (i.e., outbreak control C+ or non-outbreak control C–) in order to assign (i.e., absence of statistical differences: H0 conserved), or not (i.e., presence of statistical differences: H0 rejected), this tested genome to the outbreak of interest. The SNPs were identified by the workflow ‘iVARCall2’ against the reference genome S. Typhimurium LT2 (NCBI NC_003197.1). The produced pseudogenomes (4,857,432 bp) were inferred using the program ‘RaxML’ based on a bootstrap analysis and search for best-scoring Maximum Likelihood tree with General Time-Reversible model of substitution and the secondary structure 16-state model. Bootstraps higher than 80% are represented at each node.