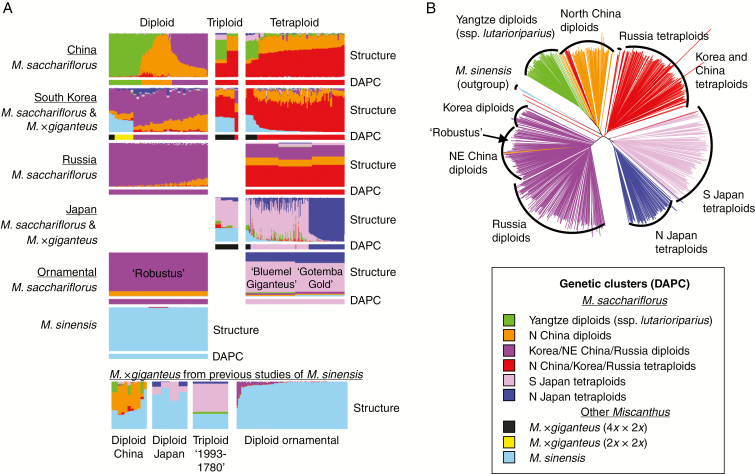
Fig. 2.
Individual-based analysis of population structure of Miscanthus sacchariflorus and M. × giganteus using RAD-seq SNP data. Individuals that had ≥20 % M. sinensis admixed with M. sacchariflorus were considered to be M. × giganteus. (A) Bar charts showing Q values from Structure analysis (top of each set by provenance) or assignment to groups based on discriminant analysis of principal components (DAPC, bottom of each set) using 34 605 RAD-seq SNPs. Shown are nine DAPC groups, comprising six M. sacchariflorus, one M. sinensis and two M. ×giganteus; in the Structure analysis, seven groups are shown with colour as for DAPC groupings except that the M. × giganteus individuals are represented as an admixture of M. sinensis and M. sacchariflorus. All 764 individuals from the current study are plotted. The bottom portion includes an additional 93 individuals from previous studies (Clark et al., 2014, 2015). ‘Ornamental’ indicates cultivars available from the horticultural nursery trade in North America. (B) Neighbor–Joining tree of 722 Miscanthus sacchariflorus individuals using 31 743 RAD-seq SNPs. Five M. sinensis individuals were included as an outgroup. Branches are coloured based on DAPC groupings (A). To reduce the signal of hybridization, 37 early generation hybrids were excluded, as were 2862 SNPs that were highly differentiated between M. sinensis and M. sacchariflorus.