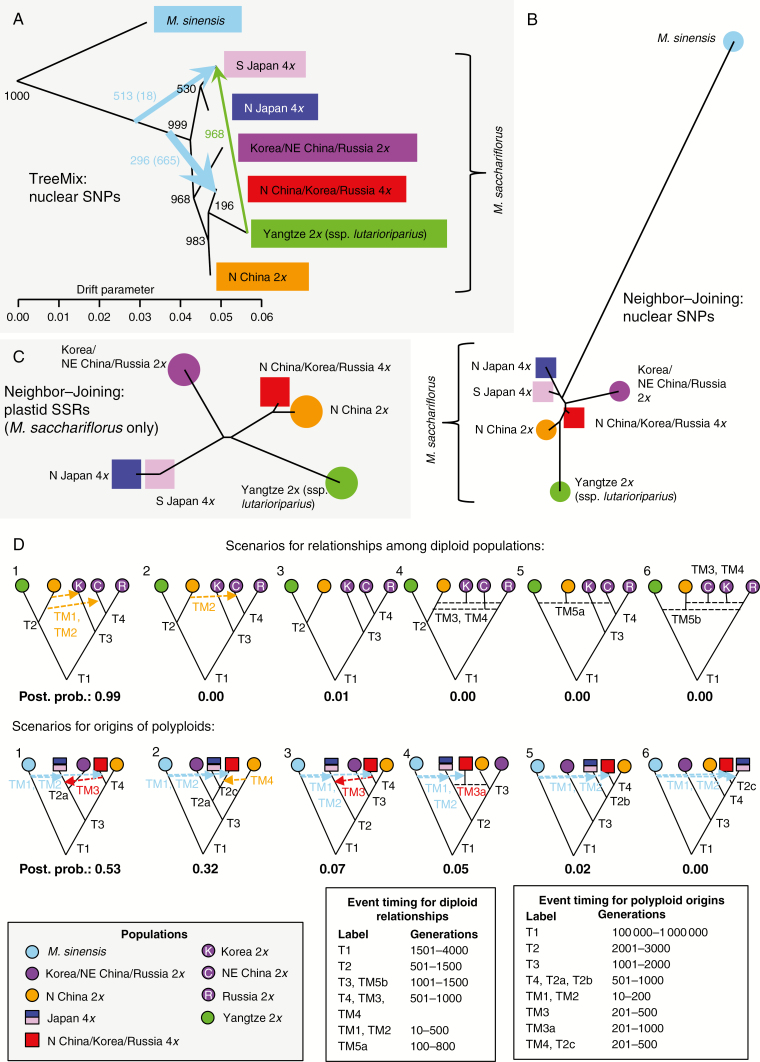
Fig. 3.
Relationships among genetic groups of Miscanthus sacchariflorus based on RAD-seq SNPs and plastid microsatellites. Groups were identified using discriminant analysis of principal components (DAPC; Fig. 2A). (A) Relationships among six M. sacchariflorus groups, and one M. sinensis outgroup, as determined by the software TreeMix based on 34 605 RAD-seq SNPs and 727 individuals. The black tree indicates population divergence. Coloured arrows indicate subsequent migration, colour coded by population of origin. Arrow width signifies the magnitude of migration. The numbers of bootstrap replicates out of 1000 supporting each clade or migration event are indicated. Numbers in parentheses indicate the number of replicates in which there was a migration event from M. sinensis to the indicated population. (B) Neighbor–Joining tree calculated from pairwise Jost’s D statistics using allele frequencies at 34 605 RAD-seq SNPs across 727 individuals. (C) Neighbor–Joining tree calculated from pairwise Jost’s D statistics using frequencies of plastid haplotypes across ten microsatellite markers, across 712 M. sacchariflorus individuals with complete plastid data (M. sinensis is not shown due to the lack of shared haplotypes with M sacchariflorus). (D) Scenarios tested by DIYABC and their estimated posterior probabilities. Arrows indicate migration events, and dashed horizontal lines indicate admixture events. Model constraints upon divergence (T) and migration times (TM) are indicated in terms of number of generations before the present.