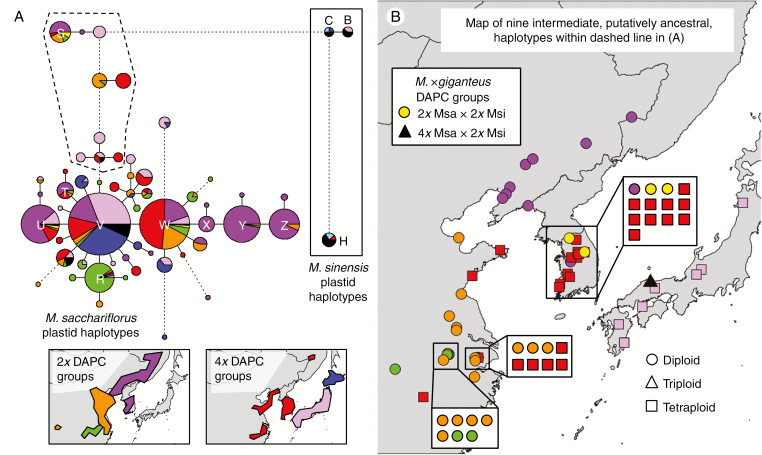
Fig. 4.
Plastid haplotypes for 759 Miscanthus spp. individuals using ten microsatellite markers. Colours from Fig. 2 indicate discriminant analysis of principal components (DAPC) groups identified with nuclear SNPs, with six M. sacchariflorus, one M. sinensis (light blue) and two M. × giganteus (yellow and black) shown; geographical distributions of M. sacchariflorus groups are shown in the inset. (A) Haplotype network. Each circle or pie chart represents one haplotype, and the area is proportional to the number of individuals with the haplotype. Letters indicate haplotypes referred to in the text. Solid lines connecting circles and pie charts indicate single mutational steps (one nucleotide difference in amplicon size at one microsatellite marker). Dotted lines indicate multiple mutational steps, proportional in number to the length of the line. Nine haplotypes surrounded by a dashed line are intermediate between common M. sacchariflorus and common M. sinensis haplotypes. (B) Map of individuals possessing any of the nine intermediate haplotypes surrounded by the dashed line in (A). Shapes indicate ploidy.