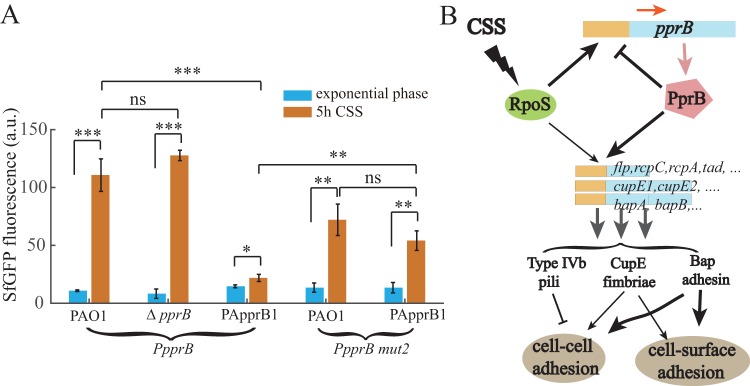
FIG 7.
PprB negatively regulates the transcription of itself, and model of CSS responses of PprB-regulated genes through RpoS. (A) Expression values of pprB or ppprB-mut2 (PprB binding sequence GGCTAATAC was mutated to GGCGGGTAC) transcriptional reporters in the wild type or pprB mutant or PApprB1 (PprB was constitutively overexpressed) strains at exponential phase or after 5-h carbon deprivation. Data are from three independent experiments and shown as the means ± SDs. Statistical analysis was based on pairwise strain comparisons (t test). *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B) Schematic representation of the RpoS-PprB-Flp/CupE/Bap/Tad system and its signaling cascade in response to CSS. CSS induces the expression of PprB-regulated genes by triggering the expression of PprB. RpoS mediates the CSS signal induction of PprB transcription. Expression of CupE fimbriae (moderately) and Bap adhesin (largely) enhances bacterial CCA and CSA, the type IVb pili have a negative effect on CCA. PprB negatively regulates the transcription of itself.