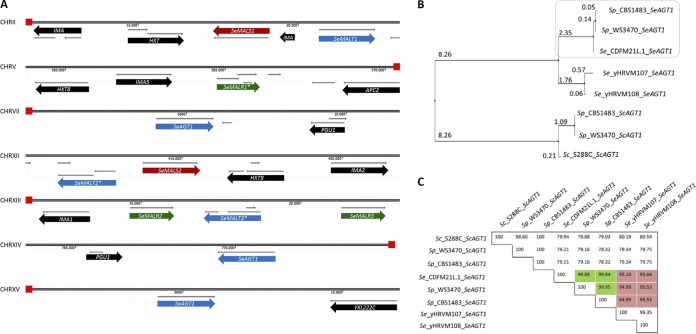
FIG 2.
Organization of subtelomeric regions involving MAL genes and SeAGT1 in CDFM21L.1. (A) Chromosome sections are represented as lines, and red boxes denote telomeres. The CDFM21L.1 genome harbors three SeMALT genes in which SeMALT2 and SeMALT3 have a mutation resulting in an early stop codon and truncated protein (denoted with *). Three copies of SeAGT1 were found close to the telomeres on chromosomes VII, XIV, and XV. Furthermore, there are two intact SeMALS genes on ChrII and ChrXII and three SeMALR genes on ChrV and ChrXIII whose copy on ChrV is also mutated (SeMALR1*). The gene and interval sizes are approximately to scale. Transporter genes SeAGT1, SeMALT1, SeMALT2, and SeMALT3 are denoted with blue arrows, the hydrolase genes SeMALS1 and SeMALS2 are denoted with red arrows, and the regulator genes SeMALR1, SeMALR2, and SeMALR3 are denoted with green arrows. Any other genes are shown with black arrows. (B) Phylogeny of Saccharomyces SeAGT1 genes described in S. cerevisiae, S. eubayanus, and the lager brewing hybrid S. pastorianus. (C) Nucleotide percentage identities between AGT1 orthologs from S. cerevisiae, S. eubayanus, and the lager brewing hybrid S. pastorianus. Green indicates highest similarity between SeAGT1 and SeAGT1 genes from S. pastorianus strains CBS 1483 and WS3470. Red indicates similarity between SeAGT1 from North American strains with SeAGT1 genes from Asian S. eubayanus and S. pastorianus strains CBS 1483 and WS3470.