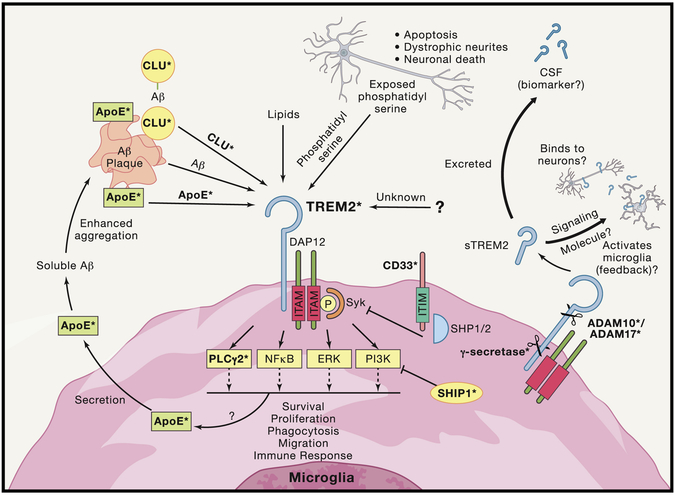
Figure 4: Convergence of Multiple AD Risk Genes/Loci on TREM2 Signaling.
TREM2 signaling pathways involve potential complex signaling interactions with other AD genetic risk factors with a diverse array of functional consequences. Studies of TREM2 have begun to uncover a number of possible ligands for TREM2 that are highly expressed in the CNS and increased in AD brain, and more ligands potentially await discovery. Among the proposed ligands for TREM2 are ApoE, Clu/ApoJ, various lipids, phosphatidyl serine, and even potentially Aβ itself. TREM2 lacks its own signal transduction component and must interact with ITAM adaptor protein DAP12 in order to mediate downstream signaling. DAP12 activation and Syk phosphorylation led to a number of potential intracellular signals including NF-kB, PI3K, ERK, and newly identified AD risk gene PLCG2. Recent evidence also indicates that activation of TREM2 increases microglial production of ApoE which may then act in reciprocal cycle binding to amyloid and then binding to TREM2. The natural inhibitor of ITAM signaling comes from ITIM signaling. Interestingly, another AD risk gene CD33 contains an ITIM signaling domain, creating a potential link between TREM2 and CD33. Additionally, the gene which encodes for the protein SHIP1 has been identified as potential risk allele in AD and studies have shown that SHIP1 inhibition of PI3K activation exerts an inhibitory effect on TREM2 signaling. Ultimately, activation of TREM2 signaling results in several potential phenotypic alterations including altered migration, survival, proliferation, cytokine release, and other functions. Which intracellular pathways are directly responsible for the different functional consequences are still being worked out and may be ligand specific. TREM2 is also cleaved into a soluble fragment (sTREM2) by α-secretase (ADAM10/17, which are also implicated in AD risk and play key roles in APP processing). The function of sTREM2 is still not fully understood but sTREM2 can be detected in CSF and is being examined as potential biomarker of disease state in AD and other CNS disorders. sTREM2 has also been recently proposed to potentially bind to neurons, as well as potentially binding to an unknown receptor reciprocally on microglial. Finally, after α-secretase cleavage the remaining membrane fragment of TREM2 is cleaved by γ-secretase (components of which, PSEN1, and PSEN2, are known to cause early onset fAD). Inhibition of this cleavage results in inability of DAP12 to release from non-functional TREM2 and therefore sequestration of DAP12. Bold terms with asterisks indicate proteins coded for by genes that alter risk of AD.