Abstract
Cystic fibrosis (CF) is caused by mutations in the CF transmembrane conductance regulator (CFTR) gene. The majority of CFTR mutations result in impaired chloride channel function as only a fraction of the mutated CFTR reaches the plasma membrane. The development of a therapeutic approach that facilitates increased cell-surface expression of CFTR could prove clinically relevant. Here, we evaluate and contrast two molecular approaches to activate CFTR expression. We find that an RNA-guided nuclease null Cas9 (dCas9) fused with a tripartite activator, VP64-p65-Rta can activate endogenous CFTR in cultured human nasal epithelial cells from CF patients. We also find that targeting BGas, a long non-coding RNA involved in transcriptionally modulating CFTR expression with a gapmer, induced both strong knockdown of BGas and concordant activation of CFTR. Notably, the gapmer can be delivered to target cells when generated as electrostatic particles with recombinant HIV-Tat cell penetrating peptide (CPP), when packaged into exosomes, or when loaded into lipid nanoparticles (LNPs). Treatment of patient-derived human nasal epithelial cells containing F508del with gapmer-CPP, gapmer-exosomes, or LNPs resulted in increased expression and function of CFTR. Collectively, these observations suggest that CRISPR/dCas-VPR (CRISPR) and BGas-gapmer approaches can target and specifically activate CFTR.
Keywords: CFTR, cystic fibrosis, lncRNA BGas, transcriptional regulation, exosome, ASO delivery, Tat-CPP
In this issue, Villamizar et al. develop a new therapeutic “gapmer” that targets and inhibits a regulator of the CF transmembrane conductance regulator (CFTR) gene. Utilizing various delivery methods, they demonstrate delivery of the gapmer to patient samples and establish a platform to advance next-generation therapies to treat cystic fibrosis.
Introduction
Cystic fibrosis (CF) is an inherited and life-limiting disease caused by mutations in the CF transmembrane conductance regulator (CFTR) gene.1 CFTR protein is expressed in specialized epithelial cells in organs such as the pancreas, sweat glands, genital ducts, and lungs. In the respiratory epithelium, CFTR functions as a cAMP-activated plasma membrane channel helping to exchange chloride ions and hydrate the surface of the lungs.2 Defects in this protein result in the accumulation of thick mucus, inflammation, tissue fibrosis, and impaired lung function.3 To date, more than 2,000 CF-associated mutations have been identified. These CFTR mutations have been classified based on the mechanisms that affect the synthesis, trafficking, and/or function of the protein. Six classes of mutations are described: class I results from non-sense or frameshift of mRNA leading to reduced protein expression, class II mutation produces a defect of the protein processing with lack of the channel opening, class III involves a defect on the channel gating, class IV generates misshaped channels causing reduced conductance function, class V mutations are characterized by splicing defects that produce low levels of CFTR protein, and class VI mutation produces a functional but relatively unstable protein at the cell surface.4
The most prevalent mutation, which affects more than 70% of patients, are the class II mutations caused by the deletion of a phenylalanine residue at position 508 (F508del) of the CFTR protein.5 The F508del protein exhibits a folding defect that, following recruitment to the endoplasmic reticulum, results in enhanced proteolytic degradation.6, 7 However, a small proportion of these misfolded proteins are released to the plasma membrane where they display a reduction in channel gating.8 Small molecules have recently been developed to assist in restoring CFTR folding and channel function.9 These small molecules, VX-809 (lumacaftor) and VX-770 (ivacaftor), enhance cellular chaperone function and act as potentiator to increase channel gating and conductance, respectively.10 Notably, the combination of the potentiator and a corrector (ivacaftor and lumacaftor), known collectively as Orkambi, was recently approved by US Food Drug Administration (FDA) for the treatment of patients with F508del mutation.11, 12
CFTR is a large gene (189 kb) encompassing 27 exons located at chromosome 7q31.2 and is transcriptionally regulated by both enhancers and cis-elements acting at the promoter and elsewhere in the locus.13 Interactions of regulatory elements in introns 1 and 11 have been shown to modulate the expression of the CFTR gene by recruiting RNA polymerase II (RNAP-II) to the promoter.14 Recently, an antisense long non-coding RNA (lncRNA) named BGas (CFTR-AS1 [ENST00000441019.1], HGNC:40144) was identified and found to modulate the expression.15 BGas was found to regulate CFTR expression by recruiting HMGB1 protein to the BGas region of the CFTR locus, which results in a localized distortion of chromatin structure and disruption of RNAP-II activity, ultimately leading to a reduction of CFTR expression.15 The role of each of these individual elements maintains the expression of CFTR and opens the potential for new epigenetic therapies. Here, we report the transcriptional activation of CFTR in human nasal epithelial cells from patients with F508del treated with either CRISP/Cas9-VPR (VP64-p65-Rta) or antisense oligonucleotide (gapmer) directed to BGas.
Gene therapy for CF has been explored in various clinical trials with little success. Early studies used adeno-associated virus (AAV) bearing the CFTR cDNA. These studies demonstrated the ability to correct chloride transport.16 However, the induction of an immune response to AAV prevented efficacy and limited the implementation of this therapy. To date, lentiviral vectors have not been tested in CF patients. One reason is that the vector integration into the host genome can cause mutagenesis.17 Non-viral vector alternatives have also been evaluated in patients with CF. In one study, cationic liposomes were used to deliver the CFTR gene. Unfortunately, only a modest improvement in lung function was observed after a year of monthly dosing of patients. This suggested that these specific liposome-delivered formulations cannot maintain CFTR expression for a prolonged period.18
One of the main challenges in the treatment of CFTR is the delivery of therapeutically efficacious moieties to the lungs. To overcome this issue, we explore three distinct approaches to deliver the CFTR activating BGas-gapmer into CF-patient-derived F508del containing cultured nasal cells. We show here that an electrostatic particle, binding gapmer with human immunodeficiency virus cell penetrating peptide Tat (HIV-Tat), and packaging the gapmer into exosomes results in a bona fide means to deliver functional antisense oligonucleotides that can activate CFTR expression. We also find that lipid nanoparticles (LNPs) are inefficient delivery vehicles for and can only modestly increase CFTR expression. Collectively, those approaches outlined here may represent new therapeutic strategies for bolstering CFTR expression that can be used in CF patients bearing common F508del mutations or other mutations that do not respond to current therapies.
Results
CFTR Transcriptional Activation by dCas9-VPR
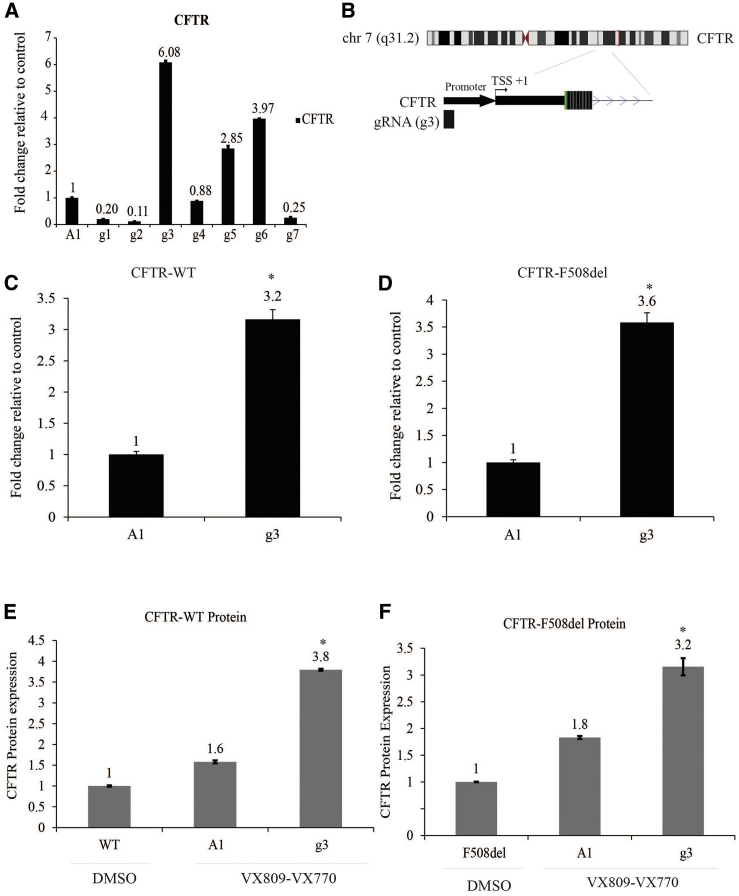
The CRISPR/Cas9 has been modified to lack endonucleolytic activity, resulting in a deactivated Cas9 (dCas9) that retains the capacity to interact with DNA. Different dCas9 systems fused with transcriptional activator domains such as VP64 (herpes virus transcription factor), p65 (nuclear factor кB [NF-кB] subunit), and Rta (an activator of Epstein-Barr virus genes) to increase the expression of those genes targeted by the small guide RNA (gRNA) have been developed.19, 20, 21 We sought to assess the ability to utilize dCas9/VPR to activate endogenous CFTR expression. We screened a total of seven gRNAs (designated g1–g7) directed to the CFTR promoter. When compared with the non-specific gRNA control (gRNA-A1), three of the seven small guide RNAs (sgRNAs) (g3, g5, and g6) demonstrated significant activation of CFTR expression (Figure 1A).
Figure 1.
Activation of CFTR Expression by dCas9/VPR
(A) Real-time qPCR of CFTR expression after screening seven gRNAs (g1–g7) directed to the CFTR promoter (A1, control) using CFPAC cells. (B) Top, chromosome 7 ideogram showing CFTR gene. Bottom, close up snapshot of the UCSC browser of the gRNA (g3) located upstream of CFTR promoter. Position is relative to the transcription start site (TSS). (C and D) CFTR promoter-directed guide RNA (g3) (C) activates CFTR mRNA expression in nasal cells from healthy donor-WT and (D) in nasal cells from a CF patient with F508del mutation. (E and F) CFTR protein overexpression enriches after treatment with dCas9/VPR. CFTR ELISA showing CFTR protein level expression increased after being treated with g3RNA/dcas9-VRP in (E) nasal cells from healthy donor-WT and (F) in nasal cells from a CF patient with F508del mutation. Experiments were performed in triplicate in cells shown with the SEMs and p values from a paired two-sided t test, *p = 0.001.
Further analysis indicated that g3, which is located upstream of the CFTR promoter (Figure 1B), exhibited the most significant increase in the CFTR expression when contrasted with the control gRNA-A1 in wild-type (WT) and F508del human nasal cells (Figures 1C and 1D). Similar observations of g3 activation of CFTR expression were also noted in the CF pancreatic adenocarcinoma (CFPAC) cells (Figure S1A). These data suggest that g3 is a good candidate for targeting dCas-VPR to transcriptionally activate CFTR expression. The CFTR protein is an ATP-binding cassette (ABC) transporter with a specific cytoplasmic regulator domain (R) whose phosphorylation by protein kinase A (PKA) activates the CFTR chloride conductance channel. Here, we evaluated CFTR protein expression after the transcriptional activation of CFTR with g3RNA/dCas9-VPR quantitatively using ELISA. Notably, F508del cells incubated with VX-809 and VX-770 enhanced the levels of CFTR protein relative to the control (cells incubated with DMSO-vehicle) (Figures 1E and 1F). These data suggest that g3RNA-dCas9/VPR ribonucleoprotein complexes (RNPs) functionally increase CFTR protein expression in the presence of VX-809 and VX-770. Notably, transient transfection of dCas9/VPR into WT CFTR-expressing cells exhibited an increased expression of the CFTR mature form band C, as determined by western blot (Figures S2A and S2B).
Gapmer-Directed Downregulation of BGas Enhances CFTR Expression
Previous studies have shown BGas as a nuclear-localized lncRNA transcribed in antisense orientation from intron 11 of the CFTR gene.15 BGas is 3,944 nucleotides in length and consists of two exons. BGas function by modulating the local DNA structure interacting with chromatin remodeling proteins HMGA1, HMGB1, and WIBG, which ultimately disrupt RNAP-II function.15 In the presence of BGas, transcription of CFTR decreases resulting in a reduction of chloride ion channel function.
Antisense oligonucleotides (ASOs) are single-stranded DNA that are typically of 13–50 nucleotides in length. ASOs bind a complementary RNA target. The formation of this DNA-RNA duplex serves as a substrate for cleavage by RNase H. RNase H, an enzyme endogenously localized in the nucleus, can be used to target nuclear localized non-coding transcripts. The phosphodiester backbone of the DNA is susceptible to nuclease degradation. To prevent the ASO degradation, chemical modifications such as phosphorothioate (PS), have been instilled in the backbone of the DNA to provide enhanced biostability and pharmacokinetics.22, 23 Here, we designed an ASO as a gapmer with PS-modified backbone surrounding an unmodified DNA-like gap. In this study, the gapmer, as well as its corresponding scramble gapmer sequences, are 20 nucleotides in length with three PS bonds at both the 5′ and the 3′ ends.
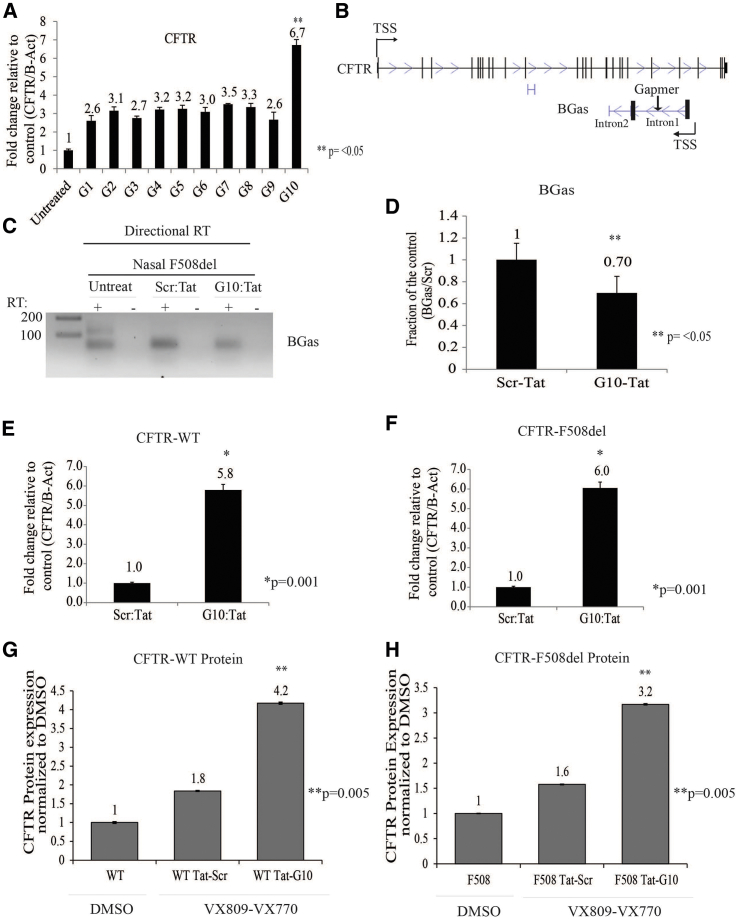
To determine the best gapmer candidate for targeting BGas, we screened 10 ASOs (designated as G1–G10) (Figure 2A). Interestingly, only gapmer 10 (G10), which is targeted to BGas intron 1 (Figure 2B), was found to activate CFTR expression (Figure 2A). Notably, G10 treatment of F508del human nasal cells resulted in a concomitant repression of BGas (Figures 2C and 2D). G10 was also found to be functional in F508del-CFTR mutation containing CFPAC cells (Figure S3A). Collectively, these data suggest that G10 is an excellent candidate for targeting the activation of CFTR by inhibiting the negative regulatory BGas lncRNA.
Figure 2.
BGas Repression Results in Increased Expression of CFTR
(A) Screen panel of ten gapmers. (B) Top, diagram of CFTR and BGas showing gapmer targeting BGas intron 1, bottom. (C) Directional RT-PCR (inverted picture of an end-point PCR) of BGas and (D) relative densitometric analysis of the individual band in nasal cells from a CF patient with F508del. (E and F) Gapmer-Tat CPP increased CFTR expression in (E) nasal cells from healthy donor-WT and (F) in nasal cells from a CF patient with F508del mutation. CFTR expression was determined by qRT-PCR. (G and H) CFTR ELISA showing CFTR protein level expression increased after being treated with BGas-gapmer in (G) nasal cells from healthy donor-WT and (H) nasal cells from a CF patient with F508del mutation. For directional RT-real-time qPCR (C) and ELISA (G and H), experiments were performed in triplicate in cells shown with the SEMs and p values from a paired two-sided t test, *p = 0.001, **p < 0.05.
Delivery is the key issue to developing virtually any genetic therapy. To overcome the issue of delivery, we generated gapmers fused with HIV-Tat cell-penetrating peptide (CPP). Tat-peptide enters cells and transit proteins across the blood-brain barrier.24 Cationic CPPs form complexes efficiently with negatively charged oligonucleotides.25 To assess the delivery of G10 to target cells, we generated a complex formation between G10 and recombinant Tat peptide through electrostatic binding. We performed a dose titration of the G10 and the Tat-CPP based on molar ratios to determine the optimal dose for efficacy (Figure S3B). We found that the molar ratio of cargo to the peptide (G10/Tat) of 10/200 μM produced the greatest increase in CFTR transcription. G10-Tat treatment of WT and F508del human nasal cells (Figures 2E and 2F) and CFPAC cells (Figure S3C) resulted in a significant increase in CFTR expression. Our findings suggest that ASOs formulated with the CPP can be utilized to directly target cells and affect CFTR expression.
Application of ASO CPP delivery has limitations such as toxicity and the induction of immune responses through Toll-like receptors (TLRs).26 In an attempt to evaluate the induction of inflammatory cytokines in nasal WT and F508del cells treated with Scramble or G10, we examined the production of interleukin-6 (IL-6). Notably, we found no differential increased expression of IL-6 compared to the high expression obtained from lipopolysaccharide (LPS)-treated cells as a positive control (Figures S4A and S4B). We also evaluated the cellular cytotoxicity of the Tat-CPP by the production of lactate dehydrogenase (LDH). A modest increase in the LDH production compared with the control (LPS) (Figure S4C). Collectively these data indicate that Tat-CPP is a relatively non-immunogenic peptide that may prove therapeutically relevant.
Gapmer-Loaded Exosomes and LNPs Increase CFTR Expression
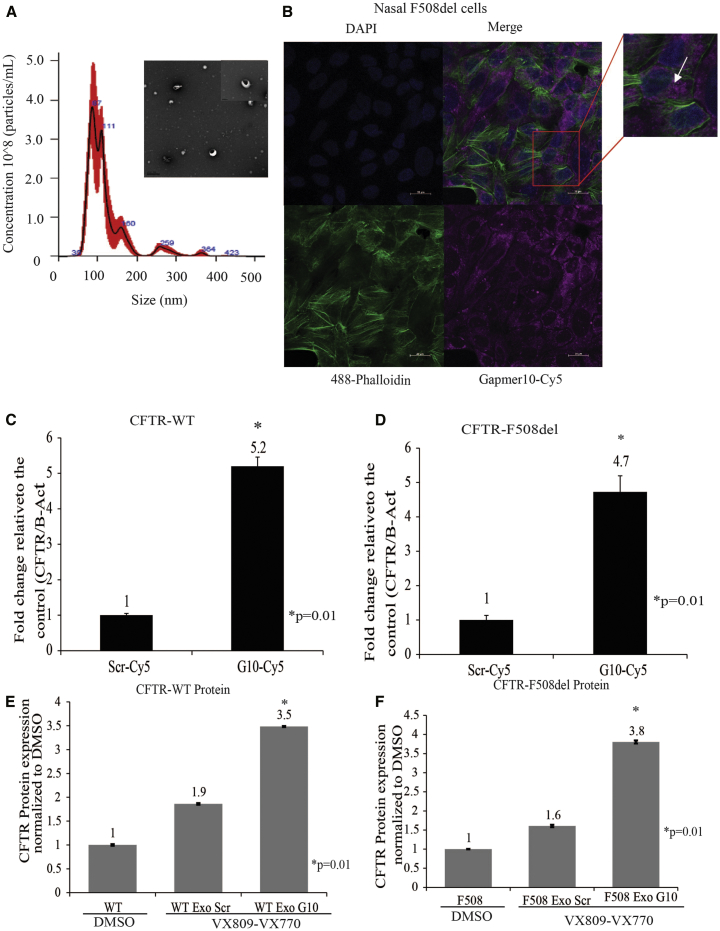
While it is relatively straightforward to affect CFTR expression by direct transfection of cells with the various biological agents, delivery in vivo to those cells requiring activation of CFTR will necessitate a more creative approach. For that reason, we decided to explore the delivery of G10 using either exosomes or LNPs. Exosomes have been found to function as vehicles to deliver protein, DNA, and/or RNA cargo.27 LNPs, similar to exosomes, have been utilized to deliver oligonucleotides to intracellular sites both in vivo and in vitro.28, 29 We sought to determine if exosomes could be used to deliver G10 to target BGas in cells. Exosomes were isolated from A549 cells and characterized for their size using nanoparticle tracking analysis (NTA) (Figure 3A). The NTA analysis demonstrated that the isolated exosomes were relatively homogeneous with an average particle size of 100 nm (Figure 3A). The expression of exosome marker CD9 was found to be present as determined by ELISA (data not shown). We also evaluated the production of LNPs (size, ∼90–110 nm) developed with a modified mixture of cationic lipids and polyethylene glycol (PEG) (described in Wu et al.28) (Figures S5A and S5B). We observed that these LNPs were functional in delivering Cy5-G10 to target cells and inhibit BGas lncRNA expression (Figures S5D and S5E).
Figure 3.
Exosome-Mediated Delivery of Gapmer Enters Human Nasal Cells and Increases Expression of CFTR
(A) Transmission electron microscopy (TEM) micrographs of exosomes isolated from the culture medium of A549 cells. Exosomes were measured by using a Nanosight NS-300 system in the supernatant from culture cells. The histogram represents particle size distribution. (B) Confocal analysis of uptake of gapmer10-Cy5 electroporated exosomes into nasal cells. (C) Real-time qPCR of CFTR mRNA levels in nasal cells from healthy donor-WT and (D) nasal cells from a CF patient with F508del mutation treated with exosome package gapmer 10 (G10) or scramble (Scr). (E and F) CFTR ELISA was carried out for CFTR protein expression in human nasal cells treated with exosomes carrying BGas-gapmer 10 (G10) or control scramble (Scr) from (E) healthy donor-WT and (F) nasal cells from a CF patient with F508del mutation. Scale bar, 20 μm. Experiments were performed in triplicate in cells shown with the SEMs and p values from a paired two-sided t test, *p = 0.001, **p < 0.05.
Next, we sought to assess the efficacy of exosome- and LNP-mediated delivery of G10 to CFTR-deficient cells from patients with CF. Cy5-labeled G10 efficiently internalized into the F508del cells (Figures 3B and S5C). To determine to what extent the exosome- and the LNP-delivered G10 was functional in activating CFTR, we assessed CFTR mRNA expression in Nasal cells WT and F508del. We observed a notable increase in CFTR expression in cells treated with the G10 exosomes relative to the scrambled controls (Figures 3C and 3D). However, in those cells treated with LNP-delivered G10, we observed only a modest increase in CFTR expression (Figures S5D and S5E). Collectively, these data suggest that G10 can be efficiently packed into exosomes and that these exosomes can be used to target cells and activate CFTR expression. Notably, these data also suggest that LNPs are relatively ineffective moieties for delivering functional G10 to target cells.
We established the functional relevance of exosome- and LNP-mediated G10 on the correction of F508del CFTR protein using ELISA. Notably, exosome-G10 treatment improved membrane localization of the mutant protein when co-administered with the potentiator (VX770) and the corrector (VX809), drugs used in combination to currently treat CF-F508del patients.10, 12 (Figures 3E and 3F). Conversely, levels of F508del CFTR protein were only modestly resolved in those cells treated with LNP/G10 and VX770 and VX809 (Figures S5F and S5G). Together, these observations demonstrate a correlation between the expression of CFTR transcript and the translated protein in vitro and provide important insights into novel methods to deliver therapeutically relevant oligonucleotides to treat CF patients.
Functional Enhanced CFTR-Mediated Halide Transport
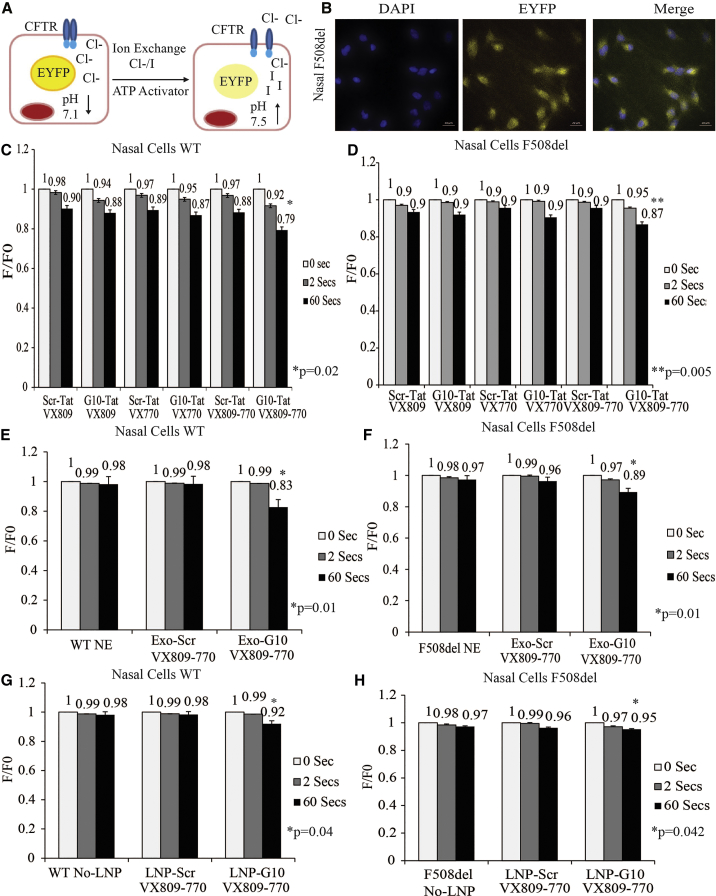
To further investigate the effect of dCas9-VPR and G10 in the trafficking of F508del to the plasma membrane and the subsequent increase in chloride transport, we capitalized on the iodide-sensitive YFP30 to measure CFTR function. Forskolin stimulation of CFTR in cells expressing a mutant form of a yellow fluorescent protein (YFP-H148Q) was assessed by measuring decreased rates of fluorescence.30 This assay is dependent on the replacement of chloride by iodide upon CFTR activation (Figure 4A) in human nasal cells (Figure 4B) and CFPAC (data not shown) cultured on 96-well plates and assayed using the fluorescence plate reader.
Figure 4.
CFTR-Mediated Halide Transport in EYFP
(A) Schematic diagram depicting the halide assay. (B) Human nasal F508del expressing EYFP. Cells were incubated with 0.3 μM of forskolin, 10 μM amiloride, and 100 μM of niflumic acid. Fluorescence measured in response to exchange of 25 mM of sodium iodide. Fluorescence decrease in human nasal cells, WT (C) and F508del (D), treated with electrostatic particle gapmer-Tat (G10-Tat) or scramble (Scr-Tat). Fluorescence change in nasal cells from healthy donor-WT (E) and nasal cells from a CF patient with F508del mutation (F) treated with no exosomes (NE) or with exosomes packed with gapmer (Exo-G10) or scramble (Exo-Scr). Functional halide assay in nasal cells from healthy donor-WT (G) and nasal cells from a CF patient with F508del mutation (H) treated with no lipid nanoparticles (No-LNP) or with LNPs loaded with gapmer (LNP-G10) or scramble (LNP-Scr). Experiments were performed in triplicate in cells shown with the SEMs and p values from a paired two-sided t test, *p = 0.01, **p 0.05.
To measure CFTR-mediated iodide transport, cells were pre-incubated with forskolin, which increases cyclic AMP to induce PKA-mediated CFTR channel gating by activating ATP binding cassette. Human nasal cells were also treated with amilirate and niflumic acid, epithelial sodium channel (ENaC), and calcium-activated chloride channel (CaCC) inhibitors, respectively. The use of both inhibitors resulted in a reduction in sodium permeability and the secretion of chloride through these ion transport proteins.31, 32
The change in YFP fluorescence in human nasal cells was evaluated in the presence of the potentiator (VX770) and the corrector (VX809).33, 34 A notable reduction in enhanced yellow fluorescent protein (EYFP) fluorescence in both WT and F508del cells treated with G10 was observed (Figures 4C and 4D). A similar reduction in the EYFP fluorescence was also observed in CFPAC cells treated with dCa9/VPR gRNA3 (data not shown) and exosome/G10 treatment (Figures 4E and 4F). Similar to our previous observations (Figures S5D and S5E), LNP-G10-treated nasal cells did not show a significant decrease in fluorescence (Figures 4G and 4H). Taken together, these data suggest that CRISPR/dCas9-VPR directed to the CFTR promoter and exosome-mediated delivery G10 directed to BGas result in the enhancement of CFTR functionality. These data also suggest that exosome-mediated delivery of G10 is more a more effective delivery approach than LNP-mediated delivery of G10.
Discussion
A paradigm shift in clinical treatment in CF has been based on the recent success with small molecules acting as CFTR modulators.9 These molecules can potentiate the ATP-dependent channel gating and correct the cellular localization of the F508del-CFTR to the apical membrane of epithelia.10 CRISPR/Cas9 technology has emerged as a tool for genome editing. Recent studies have used this approach to correct the CFTR gene in organoids and lung epithelial cells derived from induced pluripotent stem cells (iPSCs).35, 36 While valuable, these studies presented technical limitations based on the available gRNA target sites in the CFTR locus and the inefficient insertion of the donor templates into the target locus. Moreover, the sheer number of CFTR mutations resulting in disease present significant challenges to utilizing CRISPR systems to efficiently edit the various CFTR point mutations.
The data presented here demonstrated that a dCas9-VPR and a gRNA targeted to the CFTR promoter induce a robust expression of CFTR. Although several gRNAs were evaluated, only one (gRNA 3) successfully enhanced CFTR expression in human nasal cells with F508del. Consistent with the increase of the CFTR-mRNA, we also found a significant increase in the F508del CFTR protein after treatment with gRNA3-dCas9/VPR in combination with VX-809 and VX-770. Most importantly, we observed a functional restoration of the F508del mutant protein reaching the membrane. These observations suggest that dCas9-VPR might be a useful tool for the treatment of CF. However, delivery options required to carry the gRNA3 and dCas9-VPR to specific cells will indubitably be challenging.
The major obstacle currently facing CF patients and the functional correction of CFTR is the efficacy of gene delivery. A recent study reported that an antisense oligonucleotide targeting specifically the mRNA region around the F508del-encoding deletion restored CFTR function in patients with homozygous or heterozygous mutations.37 In the data presented here, we find that G10-Tat electrostatic particles are able to functionally reduce expression of the nuclear-localized BGas lncRNA. This approach resulted in increased CFTR-mRNA expression. Consistent with the increase of this CFTR transcript, we also observed increased protein expression and enhanced functionality of CFTR.
The Tat-delivery approach utilized here has been used to previously deliver ASOs37, 38 and found to be functional in various types of cells.17 We too find that G10-Tat electrostatic particles are functional and significantly enhanced chloride transport activity of F508del CFTR alone or combination with VX-809 and VX-770. Furthermore, we did not observe any increase in immunogenicity or cytotoxicity of gapmer nor with the Tat. Collectively, these data suggest that G10-Tat modulates the expression of BGas and can be considered as a therapeutic option to treat CF. It is noteworthy that the observations presented here are similar to a recent study that targeted nuclear RNAs as a specific strategy to regulate the expression of lncRNAs.38 Comparing our results obtained with the dCas9-VPR/gRNA approach targeting CFTR promoter and BGas-gapmer, we observed that both significantly increased CFTR transcript and protein.
Delivery of genetic material to the airway represents a challenge due to the mucosal barrier of the respiratory epithelium. Recent studies have shown LNPs can deliver a chemical-modified CFTR-RNA (cm-RNA).39 Although LNP-cm-RNA provided restoration of the CFTR function, the main issues of poor cellular uptake and an inability of the LNP-cm-RNA to escape the endocytic system and reach the cytosol remained enigmatic. One method to avoid these inherent issues is to utilize LNPs or cell-derived exosomes to deliver CFTR modulating agents, such as G10-Tat. Exosomes are an attractive delivery agent, as they are endowed with advantages including the inherent capacity to pass through biological membranes with reduced cytotoxicity and ultimately not be recognized as foreign.40, 41 Indeed, exosomes have been used to deliver exogenous CFTR glycoprotein and mRNA resulting in the correction of CFTR function.27 The observations presented here also demonstrate that exosomes from human lung cell lines can be utilized to deliver the CFTR modulatory BGas-gapmer. Notably, exosome-mediated delivery of gapmer rescued CFTR function in nasal cells with F508del mutation by increasing the levels of CFTR protein, suggesting that such an approach could be expanded upon by using patient-derived exosomes to tailor the targeting to those cells requiring enhanced CFTR expression while limiting inherent off-target effects.
We contrasted exosome-mediated delivery with LNPs. While we observed that G10 combined with nanoparticle encapsulation modestly increased CFTR expression, it was not as robust as exosome-mediated delivery approaches. Despite the enhanced encapsulation efficiency, LNP-mediated delivery did not increase F508del protein levels high enough to rescue CFTR function, while the exosome-mediated delivery approach did. Although these findings are encouraging, one factor is likely to have influenced this difference. Endosome release of the BGas-gapmer might be limited in the LNP-treated cells, possibly due to the retention of LNP inside late endosomes/lysosomes.42 But this eventuality remains to be determined. Collectively, the data presented here suggests that for LNP delivery to be efficient, it will need to be developed with particles that can escape endocytosis pathways. Undoubtedly, such a delivery method will require substantial optimization, in contrast to the exosome-mediated delivery approach presented here, which, while excellent with oligonucleotides, may also have its own set of challenges. One challenge with the exosome delivery approach may prove to be difficulties in packaging larger complexes, such as the G3-dCas9/VPR RNP described here into exosomes. LNPs and the conjugation of CFTR activating RNPs with Tat-CPP may also represent a significant challenge. Nevertheless, such approaches represent the next generation of cellular therapeutics emerging to treat CF.
Materials and Methods
Cell Culture
CFPAC cells,43 a ductal adenocarcinoma cell line derived from a patient with CF (ATCC Number CRL-1918) that has the most common form of the CF mutation (genotype, CFTR F508del/F508del [CF]) and exhibits ion transport activities consistent with CFTR, were maintained in Iscove’s DMEM (IDMEM) (Mediatech; Manassas, VA, USA) supplemented with 10% fetal bovine serum (Life Technologies; Carlsbad, CA, USA) and 50 μg/mL penicillin-streptomycin (pen/strep; Mediatech; Houston, TX, USA) at 37°C and 5% CO2. Nasal epithelial cells from healthy volunteers or CF patients were collected at University of Sydney Children's Hospital, Randwick, Sydney, Australia (HREC/16/SCHN/1230) and conditionaly reprogrammed and biobanked. The nasal epithelial cells (WT genotype, CFTR wt/wt [normal]), and nasal epithelial cells of CF patients (genotype, CFTR F508del/F508del [CF]) were maintained in Pneumocult EX Plus medium (StemCell Technologies) at 37°C and 5% CO2.
Construction of dCas9/VPR and sgRNA Expression Plasmids and Cell Electroporation
Pairs of complementary DNA oligonucleotides encoding the variable 20-nt sgRNA targeting sequences were annealed together to generate short double-strand DNA fragments with 4-bp overhangs (Table S1). These fragments were ligated into BsmBI-digested plasmid pc-cDNA-H1-BsmBI plasmid. The dCas9 VPR construct that served as the template for the various PCR amplifications was purchased from Addgene (#63,798; Cambridge, MA, USA). To introduce dCas9-VPR/gRNA3, cells were electroporated as described.3 In brief, cells were electroporated using Neon electroporator using 1,250 V, 20 ms, and two pulses and cultured for 48 h.
Real-Time qPCR and Directional RT Analysis of Gene Expression
To determine transcript levels of CFTR or BGas, total RNA was isolated 72 h post-transfection using the Maxwell 16 LEV simplyRNA purification kit and the Maxwell 16 research instrument (Promega; Madison, WI, USA). DNase-treated RNA samples were then standardized and reverse transcribed with Quiantitec (QIAGEN) using an oligo-deoxythymine (dT)/random nonamer primer mix or with strand-specific primers (Table S1). Real-time qPCR was carried out using Kapa Sybr Fast universal qPCR mix (Kapa Biosystems; Wilmington, MA, USA) on an Eppendorf Mastercycler realplex. Thermal cycling parameters started with 3 min at 95°C, followed by 40 cycles of 95°C for 3 s and 60°C for 30 s. To determine changes in BGas expression, directional reverse transcription (RT) was carried out using primer BGasDRT or without RT (background control). The fold change in gene expression was calculated using the 2-DDCt method. Following the RT step, real-time qPCR was carried out with primer BGasF and BGasR (Table S1). For analysis of other genes, random RT primed cDNAs were assessed for particular gene expression of IL-6 relative to β-actin using locus-specific primers (Table S1). The fold change in gene expression was calculated using the 2-DDCt method.
CFTR Immunoprecipitation (IP) and Western Blot
Western blot was performed as described before.44 In brief, A549 cells (10 × 10e6) were collected after being treated with gRNA3-dCas9/VPR. Flasks were rinsed twice with PBS, and then PBS was removed. RIPA was added, cells were scraped, and lysate was placed in a microcentrifuge tube and centrifuged for 3 min at 13,000 × g. The supernatant was recovered and a protein assay was performed. Next, 500 μg of soluble lysate was placed in a microcentrifuge tube. 3 μL A2-596 antibody directed against CFTR NBD2 domain (obtained from Cystic Fibrosis Foundation) was added45, 46 and rotated overnight at 4°C. 50 μL of Pierce protein A/G magnetic beads was added and rotated overnight. Immunocomplexes were centrifuged and wash pellets were washed three times with 1 mL RIPA. RIPA was removed, sample buffer was added, and the solution was incubated at 90°C for 10 min. Samples were loaded onto a 6% gel for SDS-PAGE. Minigels were transferred to nitrocellulose. Blots were probed with anti-CFTR antibody 596 directed against CFTR NBD2 domain (1:1,000).
BGas-Gapmer Electrostatic Binding with Tat CPP
Antisense oligonucleotide (ASO) sequences against BGas were designed using an algorithm previously reported.40 The BGas- and scramble-ASOs are 20-mer length, respectively. Both gapmers were purchased from IDT (San Jose, CA, USA). Antisense oligonucleotide targeting BGas intron and scramble (control) were designed with three nucleotides modified with PS at the 5′ and the 3′ end surrounding an unmodified DNA-like “gap.” The scramble-ASO has also PS-modified nucleotides surrounding an unmodified DNA-like “gap.” Electrostatic binding with Tat was performed by mixing Tat-CPP-(GRKKRRQRRR) (GeneScript, Piscataway, NJ, USA) with BGas- or scramble-gapmer to obtain a final concentration of 200:10 μM ratio, respectively (Table S1). After 2 h incubation at 37°C, electrostatic particles were dropped on cell culture. To evaluate the gapmer’s effect on BGas cells, they were incubated for 5 days at 37°C, 5% CO2 and then assessed using real-time qPCR for CFTR-mRNA or ELISA for protein.
Halide Assay
Stable cells expressing EYFP were obtained by transducing CFPAC with pLenti-EYFP plasmid and single cell sorted to select an EYFP cell clone. Human nasal cells WT and F508del expressing transiently EYFP by delivery of Premo Halide Sensor (Thermo Fisher Scientific; Waltham, MA, USA) by BacMam technology. The assay combines the YFP Venus halide sensor with a surrogate ion for chloride (iodide); upon stimulation of the chloride channel or transporter, iodide ions flow down the concentration gradient into the cells and quench YFP fluorescence upon binding; the amount of quench is directly proportional to the ion flux (chloride channel or transporter activity). Cells cultured on 96-well plates were treated with forskolin, VX809, VX770, amiloride (Alfa Aesa; Harverhill, MA, USA), and niflumic acid (Alfa Aesa; Harverhill, MA, USA), assayed after using Halide stimulus buffer (NaI 25 mM), and fluorescence evaluated in a plate reader (GloMax-Promega; Madison, WI, USA). Normalization for expression levels was performed by baseline correction (F/F0).
Isolation and Characterization of Exosomes
A549 cells were cultured for 72 h with growth media containing exosome-depleted fetal bovine serum (FBS) (GIBCO). Collected supernatant was centrifugated at 800 × g for 5 min, followed by an additional spin at 2,000 × g for 10 min; supernatant was filtered (0.2 μm) and ultracentrifuged at 100,000 × g for 2 hours at 4°C. Pelleted exosomes were resuspended in PBS and storage at −80°C. Detection of the exosomal protein marker CD9 was performed using ExoQuant (Biovision; Milpitas, CA, USA) overall exosome capture and quantification assay kit (Cell Media, Colorimetric). In brief, 100 μL of cultured media was added into the well and incubated 30 min at room temperature, washed, and then incubated with 100 μL of mouse anti-human exosome detection antibody solution (diluted in sample buffer at 1:500 dilution) for 2 h at 37°C. The plate was washed and incubated with 100 μL of streptavidin-horseradish peroxidase (HRP) antibody solution added to each well (diluted to 1:5,000 dilutions in 1× sample buffer) for 1 h at 37°C. 100 μL of substrate chromogenic solution was added to each well and incubated at room temperature in the dark for 5–10 min. The reaction was stopped by adding 100 μL of stopping solution to each well. The absorbance was read at 450 nm within and at 570 nm. The 570-nm measurements were subtracted from the measurement at absorbance 450 nm. The standard curve was used to determine the number of exosomes in an unknown sample.
Identification of Nanoparticles by NTA
NTA measurements were performed by using a NanoSight NS300 instrument. The size distribution and quantification of exosome preparations were analyzed by measuring the rate of Brownian motion with a NanoSight LM10 system (NanoSight; Wiltshire, UK) equipped with fast video capture and particle-tracking software. Isolated exosomes were diluted and injected into a NanoSight sample cubicle. The mean ± SD size distribution of exosomes was determined as well as the mean number of particles per milliliter.
Negative Staining Electron Microscopy of Extracellular Vesicles
Specimens at certain concentrations were absorbed to glow-discharged, carbon-coated 200 meshEM grids. Samples were prepared by conventional negative staining with 1% (w/v) uranyl acetate. Electron microscopy (EM) images were collected with an FEI Tecnai 12 transmission electron microscope (Thermo Fisher Scientific; Waltham, MA, USA) equipped with a LaB6 filament and operated at an acceleration voltage of 120 kV. Images were recorded with a Gatan 2 × 2 k CCD camera (Gatan; Pleasanton, CA, USA) at a magnification of 100 μm and a defocus value of ∼1.5 μm.
Exosome Labeling and Electroporation
Exosomal membrane was labeled with BODIPY TR ceramide, according to the manufacturer’s protocol (Molecular Probes/Invitrogen Life Technologies; Carlsbad, CA, USA). In brief, exosome pellets were re-suspended in 100 μL PBS and stained with 10 μmol/L BODIPY TR ceramide with 594-Alexa Fluor (red) fluorescence. Excess fluorescent dye was removed by using exosome spin columns (Life Technologies; Carlsbad, CA, USA). To pack exosomes with gapmer and Scramble ASOs, we used the protocol published by Lamichhane et al.47 Briefly, exosomes were electroporated with G10 or Scramble by using GenePulser Xcell electroporator (Bio-Rad; Hercules, CA, USA) in electroporation buffer (1.15 mM potassium phosphate [pH 7.2], 25 mM potassium chloride, 21% Optiprep). Electroporation was carried out at 400 V and 350 μF with one pulse using time constant program. Gapmer-packed exosomes were dropped on human nasal cells, and CFTR expression was evaluated after 120 h after culturing cells at 37°C, 5% CO2.
Confocal Microscopy and Imaging
Cells were cultivated fixed with BD cytofix/cytoperm fixation/permeabilization kit (cat. no. 554715 BD Biosciences; Franklin Lakes, NJ, USA) at room temperature for 20 min and then stained with fluorescent 488-phalloidin (green) for actin using the manufacturer’s recommendations (Molecular Probes/Invitrogen Life Technologies). Fluorescence images were collected on Zeiss LSM-700 confocal microscope. For the analysis of the cellular internalization of exosomes, Zeiss acquisition parameters, including exposure, focus, illumination, and Z stack projection, were controlled by Zen 2012 Imaging Software for Acquisition and Analysis.
Cytotoxicity Assay
Cell-mediated cytotoxicity was assessed using the CytoTox 96 non-radioactive cytotoxicity assay (Promega; Madison, WI, USA), according to the manufacturer’s protocol. In brief, Nasal WT and F508del cells (3.5 × 103 cells) were seeded on 96-well plates and maintained in Pneumocult EX Plus medium (StemCell Technologies) at 37°C and 5% CO2. Next, cells were treated with Tat-CPP to a final concentration of 200 μM and LPS 10 μg/mL incubated for 24 h. Then, 50 μL of supernatant was collected and mixed with 50 μL of CytoTox 96 substrate reagent (CytoTox 96 non-radioactive cytotoxicity assay kit) in each well. After 30 min incubation in the dark, 50 μL of stop solution was added to each well. Finally, the absorbance was read at 490 nm using plate reader (GloMax-Promega, Madison, WI, USA). LDH release percentage of control was calculated as (ODsample − ODcontrol)/ODcontrol] × 100%.
ELISA
Quantitative detection of human CFTR (Novus Biologicals; Centennial, CO, USA) was performed according to the manufacturer’s protocol as described previously.44, 45 In brief, the kit uses the sandwich ELISA principle. The microplate was coated with an antibody specific to human CFTR. Standards (100 μL) are added in duplicate to the plate. Nasal WT and F508del were freeze-thaw lysed by repeating this process three times. Samples lysates (100 μL) are added in triplicate to the plate. The plate was covered and incubated for 90 min at 37°C. After incubation, the liquid was removed out of each well and not washed. 100 μL of biotinylated detection antibody working solution was immediately added to each well, and the solution was covered with the plate sealer and gently mixed up and incubated for 1 h at 37°C. The solution was aspirated from each well. 350 μL of wash buffer was added to each well and soaked for 2 min, and the solution was aspirated from each well, and it was patted dry again with clean absorbent paper. This wash step was repeated three times. 100 μL of HRP conjugate working solution was added to each well, and the solution was covered with the plate sealer and incubated for 30 min at 37°C. The solution was aspirated from each well, and it was patted dry again with clean absorbent paper. This wash step was repeated five times. 90 μL of substrate reagent was added to each well, and the solution was covered with a new plate sealer and incubated for about 15 min at 37°C. The plate was protected from light, and 50 μL of stop solution was added to each well. Optical density (OD value) of each well at once was determined with a micro-plate reader set to 450 nm. Specification of the kit was as follows: sensitivity, 0.10 ng/mL; detection range, 0.16–10 ng/mL.
Preparation of Gapmer Lipids Nanoparticles
LNPs were prepared by mixing appropriate volumes of lipids in ethanol with an aqueous phase containing small interfering RNA (siRNA) duplexes, employing a Nanoessembler microfluidic device, followed by downstream processing as described before by Yanagi et al.48 In brief, for the encapsulation of siRNA, the desired amount of RNA was dissolved in water. Lipids at the desired molar ratio were dissolved in ethanol. The mol% ratio for the constituent lipids is 50% 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP; Avanti Polar Lipids; Alabaster, AL, USA), 5% DOPE (1,2-dioleoyl-sn-glycero-hosphoethanolamine) (Avanti Polar Lipids; Alabaster, AL, USA), 35% cholesterol (Avanti Polar Lipids), and 10% C16 PEG2000 ceramide (Avanti Polar Lipids; Alabaster, AL, USA).44 At a flow ratio of 1:3 ethanol:aqueous phases, the solutions were combined in the microfluidic device (Precision Nano Systems; Vancouver, BC, Canada). The total combined flow rate was 12 mL/min per microfluidics chip. Anywhere from 1 to 4 microfluidics chips were utilized, in a custom unit for parallelization (Precision Nano Systems), allowing a variable throughput for different batch sizes. The microfluidics chips employ a herringbone micromixer for extremely quick mixing times, yielding high encapsulation and narrow particle size distribution. The mixed material was then diluted 3× with deionized water after leaving the micromixer outlet, reducing the ethanol content using Amicon Pro Affinity concentration kit GST with 30 kDa Amicon ultra-0.5 device (Millipore; Burlington, MA, USA). Encapsulation efficiency was calculated by determining unencapsulated siRNA content by measuring the fluorescence upon the addition of RiboGreen (Molecular Probes; Eugene, OR, USA) to the LNP slurry (Fi) and comparing this value to the total siRNA content obtained upon lysis of the LNPs by 1% Triton X-100 (Ft), where % encapsulation = (Ft − Fi)/Ft × 100.
Author Contributions
O.V., T.S., and K.V.M. conceived the study collaboratively. O.V. performed the majority of the experiments and statistical analysis. T.S. and N.G. designed and cloned gRNAs and dCas9-VPR. S.A.W. created WT and CF nasal epithelial cell models. R.U. performed the NTA analysis. A.D. isolated human lung exosomes. O.V. and K.V.M. wrote the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank Loren Quintanar and Brian Armstrong at Light Microscopy and Digital Imaging Core at the City of Hope. We thank Marcia Miller, Zhuo Li, and Ricardo Zerda at City of Hope Electron Microscopy Core Facility for their help with electron microscopy. Research reported in this publication included work performed in the Analytical Cytometry Core supported by the National Cancer Institute of the NIH under award number P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This project was funded by NIDDK R01 DK104681-01 to K.V.M. S.A.W. and the miCF biobank are supported by Sydney Children Hospital Foundation and Cystic Fibrosis Research Trust Australia (RG173352).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.07.002.
Supplemental Information
References
- 1.Kerem B., Rommens J.M., Buchanan J.A., Markiewicz D., Cox T.K., Chakravarti A., Buchwald M., Tsui L.C. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 2.Sheppard D.N., Welsh M.J. Structure and function of the CFTR chloride channel. Physiol. Rev. 1999;79(1 Suppl):S23–S45. doi: 10.1152/physrev.1999.79.1.S23. [DOI] [PubMed] [Google Scholar]
- 3.Dalemans W., Barbry P., Champigny G., Jallat S., Dott K., Dreyer D., Crystal R.G., Pavirani A., Lecocq J.P., Lazdunski M. Altered chloride ion channel kinetics associated with the delta F508 cystic fibrosis mutation. Nature. 1991;354:526–528. doi: 10.1038/354526a0. [DOI] [PubMed] [Google Scholar]
- 4.Veit G., Avramescu R.G., Chiang A.N., Houck S.A., Cai Z., Peters K.W., Hong J.S., Pollard H.B., Guggino W.B., Balch W.E. From CFTR biology toward combinatorial pharmacotherapy: expanded classification of cystic fibrosis mutations. Mol. Biol. Cell. 2016;27:424–433. doi: 10.1091/mbc.E14-04-0935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bobadilla J.L., Macek M., Jr., Fine J.P., Farrell P.M. Cystic fibrosis: a worldwide analysis of CFTR mutations--correlation with incidence data and application to screening. Hum. Mutat. 2002;19:575–606. doi: 10.1002/humu.10041. [DOI] [PubMed] [Google Scholar]
- 6.Du K., Sharma M., Lukacs G.L. The DeltaF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR. Nat. Struct. Mol. Biol. 2005;12:17–25. doi: 10.1038/nsmb882. [DOI] [PubMed] [Google Scholar]
- 7.Thomas P.J., Ko Y.H., Pedersen P.L. Altered protein folding may be the molecular basis of most cases of cystic fibrosis. FEBS Lett. 1992;312:7–9. doi: 10.1016/0014-5793(92)81399-7. [DOI] [PubMed] [Google Scholar]
- 8.Kim S.J., Skach W.R. Mechanisms of CFTR Folding at the Endoplasmic Reticulum. Front. Pharmacol. 2012;3:201. doi: 10.3389/fphar.2012.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mijnders M., Kleizen B., Braakman I. Correcting CFTR folding defects by small-molecule correctors to cure cystic fibrosis. Curr. Opin. Pharmacol. 2017;34:83–90. doi: 10.1016/j.coph.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 10.Van Goor F., Hadida S., Grootenhuis P.D., Burton B., Stack J.H., Straley K.S., Decker C.J., Miller M., McCartney J., Olson E.R. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc. Natl. Acad. Sci. USA. 2011;108:18843–18848. doi: 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becq F., Mall M.A., Sheppard D.N., Conese M., Zegarra-Moran O. Pharmacological therapy for cystic fibrosis: from bench to bedside. J Cyst Fibros. 2011;10(Suppl 2):S129–S145. doi: 10.1016/S1569-1993(11)60018-0. [DOI] [PubMed] [Google Scholar]
- 12.Kopeikin Z., Yuksek Z., Yang H.Y., Bompadre S.G. Combined effects of VX-770 and VX-809 on several functional abnormalities of F508del-CFTR channels. J. Cyst. Fibros. 2014;13:508–514. doi: 10.1016/j.jcf.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Moisan S., Berlivet S., Ka C., Le Gac G., Dostie J., Férec C. Analysis of long-range interactions in primary human cells identifies cooperative CFTR regulatory elements. Nucleic Acids Res. 2016;44:2564–2576. doi: 10.1093/nar/gkv1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang R., Kerschner J.L., Gosalia N., Neems D., Gorsic L.K., Safi A., Crawford G.E., Kosak S.T., Leir S.H., Harris A. Differential contribution of cis-regulatory elements to higher order chromatin structure and expression of the CFTR locus. Nucleic Acids Res. 2016;44:3082–3094. doi: 10.1093/nar/gkv1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saayman S.M., Ackley A., Burdach J., Clemson M., Gruenert D.C., Tachikawa K., Chivukula P., Weinberg M.S., Morris K.V. Long Non-coding RNA BGas Regulates the Cystic Fibrosis Transmembrane Conductance Regulator. Mol. Ther. 2016;24:1351–1357. doi: 10.1038/mt.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zabner J., Couture L.A., Gregory R.J., Graham S.M., Smith A.E., Welsh M.J. Adenovirus-mediated gene transfer transiently corrects the chloride transport defect in nasal epithelia of patients with cystic fibrosis. Cell. 1993;75:207–216. doi: 10.1016/0092-8674(93)80063-k. [DOI] [PubMed] [Google Scholar]
- 17.Marquez Loza L.I., Yuen E.C., McCray P.B., Jr. Lentiviral Vectors for the Treatment and Prevention of Cystic Fibrosis Lung Disease. Genes (Basel) 2019;10:E218. doi: 10.3390/genes10030218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alton E.W., Boyd A.C., Porteous D.J., Davies G., Davies J.C., Griesenbach U., Higgins T.E., Gill D.R., Hyde S.C., Innes J.A., UK Cystic Fibrosis Gene Therapy Consortium * A Phase I/IIa Safety and Efficacy Study of Nebulized Liposome-mediated Gene Therapy for Cystic Fibrosis Supports a Multidose Trial. Am. J. Respir. Crit. Care Med. 2015;192:1389–1392. doi: 10.1164/rccm.201506-1193LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chavez A., Scheiman J., Vora S., Pruitt B.W., Tuttle M., P R Iyer E., Lin S., Kiani S., Guzman C.D., Wiegand D.J. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeder M.L., Linder S.J., Cascio V.M., Fu Y., Ho Q.H., Joung J.K. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castanotto D., Lin M., Kowolik C., Wang L., Ren X.Q., Soifer H.S., Koch T., Hansen B.R., Oerum H., Armstrong B. A cytoplasmic pathway for gapmer antisense oligonucleotide-mediated gene silencing in mammalian cells. Nucleic Acids Res. 2015;43:9350–9361. doi: 10.1093/nar/gkv964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen X., Corey D.R. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2018;46:1584–1600. doi: 10.1093/nar/gkx1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailus B.J., Pyles B., McAlister M.M., O’Geen H., Lockwood S.H., Adams A.N., Nguyen J.T., Yu A., Berman R.F., Segal D.J. Protein Delivery of an Artificial Transcription Factor Restores Widespread Ube3a Expression in an Angelman Syndrome Mouse Brain. Mol. Ther. 2016;24:548–555. doi: 10.1038/mt.2015.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McClorey G., Banerjee S. Cell-Penetrating Peptides to Enhance Delivery of Oligonucleotide-Based Therapeutics. Biomedicines. 2018;6:E51. doi: 10.3390/biomedicines6020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agrawal S., Kandimalla E.R. Role of Toll-like receptors in antisense and siRNA [corrected] Nat. Biotechnol. 2004;22:1533–1537. doi: 10.1038/nbt1042. [DOI] [PubMed] [Google Scholar]
- 27.Vituret C., Gallay K., Confort M.P., Ftaich N., Matei C.I., Archer F., Ronfort C., Mornex J.F., Chanson M., Di Pietro A. Transfer of the Cystic Fibrosis Transmembrane Conductance Regulator to Human Cystic Fibrosis Cells Mediated by Extracellular Vesicles. Hum. Gene Ther. 2016;27:166–183. doi: 10.1089/hum.2015.144. [DOI] [PubMed] [Google Scholar]
- 28.Wu S.Y., Putral L.N., Liang M., Chang H.I., Davies N.M., McMillan N.A. Development of a novel method for formulating stable siRNA-loaded lipid particles for in vivo use. Pharm. Res. 2009;26:512–522. doi: 10.1007/s11095-008-9766-1. [DOI] [PubMed] [Google Scholar]
- 29.Clarke D., Idris A., McMillan N.A.J. Development of novel lipidic particles for siRNA delivery that are highly effective after 12 months storage. PLoS ONE. 2019;14:e0211954. doi: 10.1371/journal.pone.0211954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galietta L.J., Haggie P.M., Verkman A.S. Green fluorescent protein-based halide indicators with improved chloride and iodide affinities. FEBS Lett. 2001;499:220–224. doi: 10.1016/s0014-5793(01)02561-3. [DOI] [PubMed] [Google Scholar]
- 31.Althaus M. ENaC inhibitors and airway re-hydration in cystic fibrosis: state of the art. Curr. Mol. Pharmacol. 2013;6:3–12. doi: 10.2174/18744672112059990025. [DOI] [PubMed] [Google Scholar]
- 32.Berdiev B.K., Qadri Y.J., Benos D.J. Assessment of the CFTR and ENaC association. Mol. Biosyst. 2009;5:123–127. doi: 10.1039/b810471a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eckford P.D., Ramjeesingh M., Molinski S., Pasyk S., Dekkers J.F., Li C., Ahmadi S., Ip W., Chung T.E., Du K. VX-809 and related corrector compounds exhibit secondary activity stabilizing active F508del-CFTR after its partial rescue to the cell surface. Chem. Biol. 2014;21:666–678. doi: 10.1016/j.chembiol.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 34.Ramsey B.W., Davies J., McElvaney N.G., Tullis E., Bell S.C., Dřevínek P., Griese M., McKone E.F., Wainwright C.E., Konstan M.W., VX08-770-102 Study Group A CFTR potentiator in patients with cystic fibrosis and the G551D mutation. N. Engl. J. Med. 2011;365:1663–1672. doi: 10.1056/NEJMoa1105185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Firth A.L., Menon T., Parker G.S., Qualls S.J., Lewis B.M., Ke E., Dargitz C.T., Wright R., Khanna A., Gage F.H., Verma I.M. Functional Gene Correction for Cystic Fibrosis in Lung Epithelial Cells Generated from Patient iPSCs. Cell Rep. 2015;12:1385–1390. doi: 10.1016/j.celrep.2015.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwank G., Koo B.K., Sasselli V., Dekkers J.F., Heo I., Demircan T., Sasaki N., Boymans S., Cuppen E., van der Ent C.K. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Sermet-Gaudelus I., Clancy J.P., Nichols D.P., Nick J.A., De Boeck K., Solomon G.M., Mall M.A., Bolognese J., Bouisset F., den Hollander W. Antisense oligonucleotide eluforsen improves CFTR function in F508del cystic fibrosis. J. Cyst. Fibros. 2018;18:536–542. doi: 10.1016/j.jcf.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wheeler T.M., Leger A.J., Pandey S.K., MacLeod A.R., Nakamori M., Cheng S.H., Wentworth B.M., Bennett C.F., Thornton C.A. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson E., MacDonald K.D., Slaughter K., McKinney M., Patel S., Sun C., Sahay G. Lipid Nanoparticle-Delivered Chemically Modified mRNA Restores Chloride Secretion in Cystic Fibrosis. Mol. Ther. 2018;26:2034–2046. doi: 10.1016/j.ymthe.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ha D., Yang N., Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm. Sin. B. 2016;6:287–296. doi: 10.1016/j.apsb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horibe S., Tanahashi T., Kawauchi S., Murakami Y., Rikitake Y. Mechanism of recipient cell-dependent differences in exosome uptake. BMC Cancer. 2018;18:47. doi: 10.1186/s12885-017-3958-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahay G., Querbes W., Alabi C., Eltoukhy A., Sarkar S., Zurenko C., Karagiannis E., Love K., Chen D., Zoncu R. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 2013;31:653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McIntosh J.C., Schoumacher R.A., Tiller R.E. Pancreatic adenocarcinoma in a patient with cystic fibrosis. Am. J. Med. 1988;85:592. doi: 10.1016/s0002-9343(88)80116-5. [DOI] [PubMed] [Google Scholar]
- 44.Peters K.W., Okiyoneda T., Balch W.E., Braakman I., Brodsky J.L., Guggino W.B., Penland C.M., Pollard H.B., Sorscher E.J., Skach W.R. CFTR Folding Consortium: methods available for studies of CFTR folding and correction. Methods Mol. Biol. 2011;742:335–353. doi: 10.1007/978-1-61779-120-8_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farinha C.M., Penque D., Roxo-Rosa M., Lukacs G., Dormer R., McPherson M., Pereira M., Bot A.G., Jorna H., Willemsen R. Biochemical methods to assess CFTR expression and membrane localization. J. Cyst. Fibros. 2004;3(Suppl 2):73–77. doi: 10.1016/j.jcf.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 46.Gentzsch M., Chang X.B., Cui L., Wu Y., Ozols V.V., Choudhury A., Pagano R.E., Riordan J.R. Endocytic trafficking routes of wild type and DeltaF508 cystic fibrosis transmembrane conductance regulator. Mol. Biol. Cell. 2004;15:2684–2696. doi: 10.1091/mbc.E04-03-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamichhane T.N., Leung C.A., Douti L.Y., Jay S.M. Ethanol Induces Enhanced Vascularization Bioactivity of Endothelial Cell-Derived Extracellular Vesicles via Regulation of MicroRNAs and Long Non-Coding RNAs. Sci Rep. 2017;7 doi: 10.1038/s41598-017-14356-2. 13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yanagi T., Tachikawa K., Wilkie-Grantham R., Hishiki A., Nagai K., Toyonaga E., Chivukula P., Matsuzawa S. Lipid Nanoparticle-mediated siRNA Transfer Against PCTAIRE1/PCTK1/Cdk16 Inhibits In Vivo Cancer Growth. Mol Ther Nucleic Acids. 2016;5 doi: 10.1038/mtna.2016.40. e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.