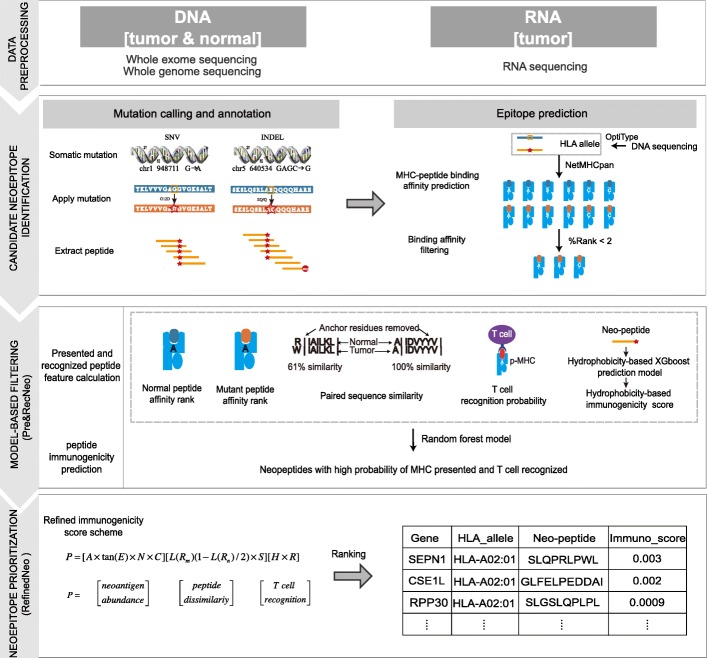
Fig. 1.
pTuneos comprises four steps: (1) Data preprocessing: raw sequencing data (WGS/WES and/or RNA-seq) are analyzed to identify somatic mutations (SNVs and indels) in VCF files and expression profiles. (2) Candidate neoantigen identification: for single nucleotide variants, a nucleotide change is translated into the corresponding amino acid change, which is then applied to the proteome reference, and nucleotide insertion and deletion changes are applied directly to the cDNA reference and translated into a 21-mer peptide containing the variant sites. The long peptide is then chopped up into 9–11-mer peptides. Peptide MHC binding affinities are then determined by NetMHCpan version 4.0 for both the mutant and normal peptides. (3) Model-based filtering: pTuneos constructs a random forest model, Pre&RecNeo, to predict the MHC presentation and T cell recognition probability of neopeptides based on five related features. (4) Neoepitope prioritization: pTuneos developed a scoring model, RefinedNeo, to refine the rank the of neoepitope immunogenicity, which represents the probability of naturally processed, MHC-presented, and T cell-recognized neopeptide and the actual immunologic effects of a neopeptide in clinical tumor treatment