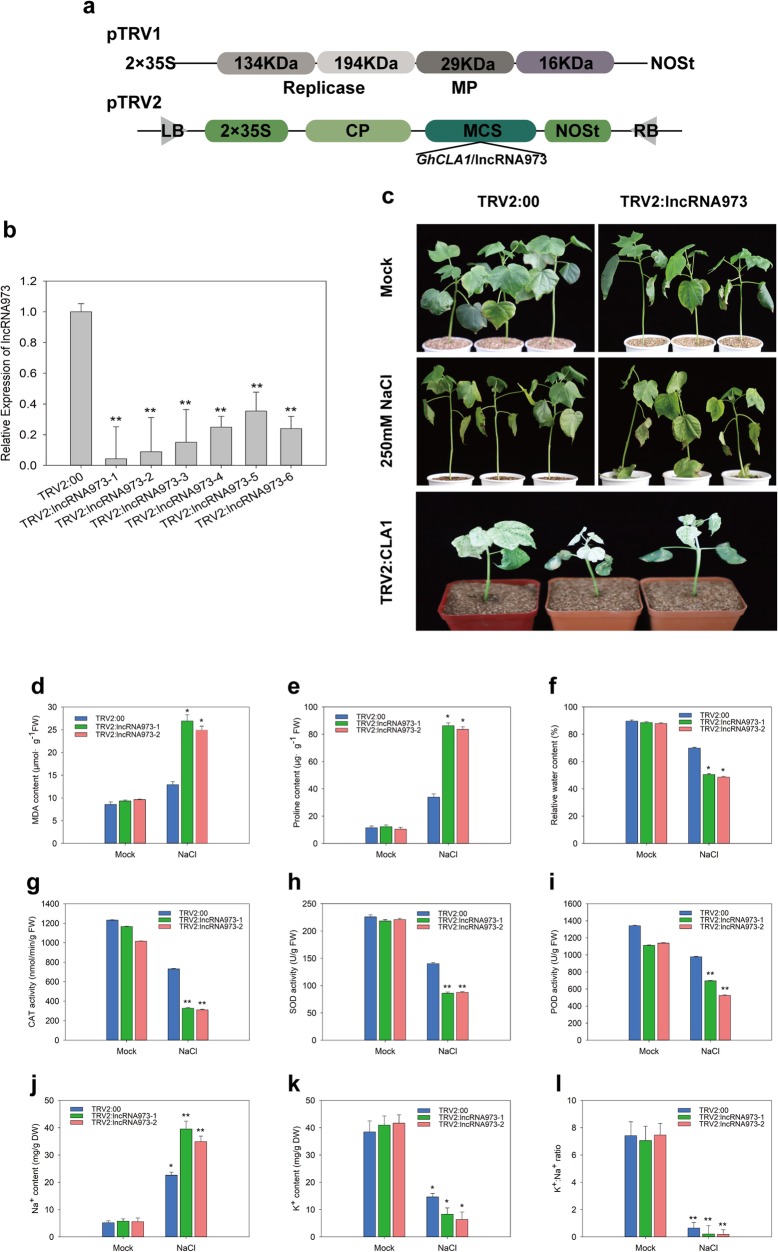
Fig. 4.
Functional identification of lncRNA973 towards salt stress using a virus-induced gene silencing (VIGS) method. a. Construction of the TRV2:GhCLA1 and lncRNA973 VIGS vectors. NOS, used as resistance selection. b. Relative lncRNA973 transcript levels in leaves of TRV2:lncRNA973 and control plants (TRV2:00). GhUBQ7 was used as an internal control. Each bar value represents mean ± SD of three independent experiments. c. Phenotypes of the silenced lncRNA973 plants at 3-d salt treatment, showing the wilting phenotype, etiolated leaves. Mock: Means to pour the same amount of water. Silencing of the endogenous Cloroplastos alterados 1 (CLA1) in cotton through TRV-mediated VIGS was used as the positive control. 10-d-old cotton seedlings (G. hirsutum L., cv. SN91–11) with two cotyledons were infiltrated with TRV2:CLA1, and plants started to display an albino phenotype in their true leaves at approximately 2 weeks later. d-f. MDA, relative water and Pro contents in the leaves under salt stress. g-i. Activities of ROS-scavenging enzymes after treatment with salt: g: catalase (CAT), h: superoxide dismutase (SOD), i: peroxidase (POD), respectively. j-l. Sodium (Na+) and potassium (K+) accumulation and K+/Na+ ratio in TRV2:00 and the TRV2:lncRNA973 plants under water and NaCl condition. Mock, with water treatment. NaCl: 250 mM NaCl treatment. *, p < 0.05 and **, p < 0.01 by Student’s t-test compared with untreated (Mock) or salt-treated TRV2 seedlings. Data represent means ± SD