Significance
Mammalian cells are constantly exposed to genotoxic agents that can lead to DNA damage, genomic instability, and diseases, including cancers. Maintenance of genomic stability, a prerequisite for survival and proper functions of cells, is facilitated by the cellular DNA repair machinery. One of the earliest responses to DNA damage is a transient inhibition of transcription to avoid fatal collisions between the DNA repair and transcriptional machineries. However, the mechanisms underlying this early transcriptional inhibition are poorly understood. Our study establishes a critical role for AFF1, a key component of super elongation complex, in early transcription inhibition and cell survival upon DNA damage, as well as a mechanism involving p300-mediated acetylation of AFF1 and consequent inactivation of the super elongation complex.
Keywords: transcription, acetylation, super elongation complex, DNA repair, p300
Abstract
Soon after exposure to genotoxic reagents, mammalian cells inhibit transcription to prevent collisions with repair machinery and to mount a proper DNA damage response. However, mechanisms underlying early transcriptional inhibition are poorly understood. In this report, we show that site-specific acetylation of super elongation complex (SEC) subunit AFF1 by p300 reduces its interaction with other SEC components and impairs P-TEFb−mediated C-terminal domain phosphorylation of RNA polymerase II both in vitro and in vivo. Reexpression of wild-type AFF1, but not an acetylation mimic mutant, restores SEC component recruitment and target gene expression in AFF1 knockdown cells. Physiologically, we show that, upon genotoxic exposure, p300-mediated AFF1 acetylation is dynamic and strongly correlated with concomitant global down-regulation of transcription—and that this can be reversed by overexpression of an acetylation-defective AFF1 mutant. Therefore, we describe a mechanism of dynamic transcriptional regulation involving p300-mediated acetylation of a key elongation factor during genotoxic stress.
Upon exposure to genotoxic stress, mammalian cells respond in a variety of ways that include initial transcriptional inhibition, cell cycle arrest, and DNA repair responses to preserve genomic integrity (1–4). Initial transcriptional inhibition is believed to be required, in part, for avoiding collision with the DNA repair machinery and for mounting a proper repair response (2, 3). Several different mechanisms have been described for transcriptional inhibition and include 1) inhibition of transcription initiation from promoters and 2) ubiquitylation of elongating RNA polymerase II (Pol II hereafter) and consequent degradation through proteolytically cleaved Def1-mediated recruitment of Elongin–Cullin E3 ligase complex (5–8), which is widely known as the “mechanism of last resort.” A role for limiting Pol II in overall transcriptional inhibition was further substantiated through in vitro transcription assays using nuclear extract of ultraviolet (UV)-irradiated cells, which showed that Pol II is a critical limiting component in overall transcriptional inhibition (9). However, initial recognition of DNA lesions by the ongoing transcriptional machinery is an important step for mounting a transcription-coupled nucleotide excision repair response for overall repair of damaged DNA. Two elegant studies have recently described a role of dose-dependent release of Pol II from promoter regions for recognition of DNA lesions and subsequent repair immediately following UV treatment (10, 11). Upon sensing DNA lesions, the elongating Pol II machinery stalls and subsequently helps in recruiting factors to repair the damaged DNA. Thus, it is quite obvious that factors associated with elongating Pol II could play an important role in overall transcriptional inhibition as well as repair-related response.
Among the many elongation factors, the recently described super elongation complex (SEC) has gained significant attention in relation to its role in transcriptional regulation. Human SEC was described as a megadalton complex containing elongation factors P-TEFb (a heterodimer of CyclinT1 and CDK9) and ELL in association with AFF1 or AFF4, AF9 or ENL, and EAF1/2 (12–15). With the exception of P-TEFb components and EAF1/2, all other SEC components are frequently fused with the N terminus of mixed lineage leukemia (MLL) to give rise to MLL fusion proteins that ultimately result in acute forms of myeloid and lymphoid leukemia (16, 17). Among all of the SEC components, the AFF1 and AFF4 paralogues are presumed to play a scaffolding role for overall SEC assembly and function. Genome-wide chromatin immunoprecipitation sequencing (ChIP-Seq) analyses have shown a significant presence of SEC components within the coding regions of target genes, suggesting that the SEC components may travel with elongating Pol II and that associated functions may have important implications for the overall regulation of transcription elongation (18, 19). Indeed, the SEC component ELL has been shown to stimulate transcription elongation by Pol II through suppression of transient pausing at pause sites within the coding region (20, 21). Therefore, it can be envisaged that the elongation activity of Pol II could also be temporally altered through modulation of SEC component activities during exposure to genotoxic stress. However, precise mechanisms of temporal regulation of transcription through SEC components are completely unknown.
A recent study has described a role for ELL in transcriptional restart after initial inhibition upon UV damage, whereas ATM-mediated phosphorylation of ENL has been shown to inhibit transcription around double-strand break (DSB) sites through recruitment of the PRC1 complex (22, 23). However, global transcriptional inhibition upon exposure to genotoxic stress through modulation of SEC activity is completely unknown. In this regard, and since AFF1 plays a scaffolding role in overall SEC assembly, it can be hypothesized that SEC assembly, and thus its function, can be regulated temporally through temporal modulation of AFF1. Consistent with this hypothesis, we report that human AFF1 is acetylated specifically by p300 both in vitro and within mammalian cells. Acetylation of AFF1 is shown to reduce its interaction with other SEC components and to reduce P-TEFb−mediated phosphorylation of the Pol II CTD in vitro, thus indicating a negative role of AFF1 acetylation in transcriptional activation. Interestingly, reexpression of wild-type (WT), but not acetylation mimic mutant, AFF1 restores SEC component recruitment and target gene expression in AFF1 knockdown cells. Finally, consistent with a negative role of AFF1 acetylation in transcriptional regulation, dynamic AFF1 acetylation is strongly correlated with transcriptional inhibition observed upon exposure of cells to genotoxic reagents.
Results
AFF1 Protein Is Specifically Acetylated by p300 within Mammalian Cells.
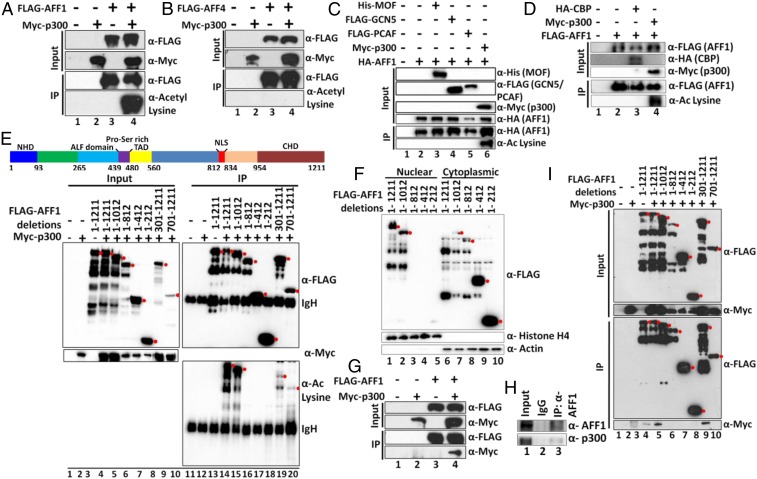
Although SEC components have been well documented with respect to functions in transcriptional regulation, temporal regulation of SEC functions for transcriptional control is unclear. To address this issue, we focused on AFF1 in view of its proposed function as a scaffolding protein for overall SEC assembly (13, 15, 24–27). Posttranslational modifications of proteins play important roles in dynamic and temporal regulation of a variety of transcription factors that include p53 (28). Since AFF1-interacting partners CyclinT1 and CDK9 have been shown to be regulated by p300-mediated acetylation (29–31), we initially tested whether AFF1 could also be acetylated by p300 within mammalian cells. Cotransfection of AFF1 and p300-expressing plasmids in 293T cells (Fig. 1A, input lanes) and subsequent immunoprecipitation and immunoblot analyses using pan-acetyl lysine-specific antibody showed a robust p300-dependent acetylation (Fig. 1A, compare lane 3 with lane 4 of IP samples). Since the AFF1 paralogue AFF4 also shows functional activity similar to that of AFF1 (15), we tested whether AFF4 might also be subject to p300-mediated acetylation. As shown in Fig. 1B, although ectopic AFF4 and p300 were well expressed (input lanes), no AFF4 acetylation was observed in this assay. Since mammalian cells contain multiple acetyl transferases that have been reported to acetylate nonhistone proteins for their functional regulation, we assessed the enzyme specificity of AFF1 acetylation in cell-based assays. As shown in Fig. 1C, among the 4 acetyl-transferases (p300, MOF, GCN5, and PCAF) that were initially tested, only p300 could acetylate AFF1 protein within mammalian cells. Interestingly, and surprisingly, a subsequent analysis also showed an inability of the p300-related CBP to acetylate AFF1 within mammalian cells (Fig. 1D), further indicating a p300-specific acetylation of AFF1.
Fig. 1.
The p300-specific acetylation of AFF1 within mammalian cells. (A) Analysis of AFF1 acetylation by p300. The 293T cells were cotransfected with indicated plasmids, and acetylation of AFF1 protein was assessed by immunoblot of anti-FLAG immunoprecipitates using pan-acetyl lysine-specific antibody. (B) Analysis of AFF4 acetylation by p300. The 293T cells were cotransfected with indicated plasmids, and acetylation of AFF4 protein was assessed by immunoblot of anti-FLAG immunoprecipitates using pan-acetyl lysine-specific antibody. (C) Analysis of the specificity of AFF1 acetylation by various acetyl transferases. The 293T cells were cotransfected with indicated plasmids, and acetylation of AFF1 protein was assessed by immunoblot of anti-FLAG immunoprecipitates using pan-acetyl lysine-specific antibody. (D) Analysis of AFF1 acetylation by CBP. The 293T cells were cotransfected with CBP- and p300-expressing plasmids, and acetylation of AFF1 protein was assessed by immunoblot of anti-FLAG immunoprecipitates using pan-acetyl lysine-specific antibody. AFF1 acetylation by p300 was used as a positive control in this experiment. (E) Analysis of AFF1-specific domains acetylated by p300. The 293T cells were cotransfected with indicated plasmids containing various AFF1 domains and p300. Acetylation was monitored as described above. The topmost band (marked by red dot) indicates the target protein, while the bottom bands are degradation products. (F) Analysis of the subcellular localization of AFF1 domains. The 293T cells were transfected with indicated AFF1 domain expressing plasmids. Nuclear and cytoplasmic fractions were prepared, and the presence of individual AFF1 fragments was analyzed by immunoblot with anti-FLAG antibody. The topmost band (marked by red dot) indicates the target protein, while the bottom bands are degradation products. (G) Analysis of an intracellular interaction of ectopic AFF1 and p300 proteins. The 293T cells were cotransfected with AFF1- and p300-expressing plasmids, and anti-FLAG immunoprecipitates were immunoblotted with anti-FLAG and anti-MYC antibodies. (H) Analysis of an intracellular interaction of endogenous AFF1 and p300 proteins. Endogenous AFF1 was immunoprecipitated using AFF1-specific antibody, and p300 was detected by immunoblotting using p300-specific antibody. (I) Analysis of p300-interacting AFF1 domains. The 293T cells were cotransfected with plasmids expressing p300 and indicated AFF1 fragments. Anti-FLAG immunoprecipitates were immunoblotted with anti-FLAG and anti-MYC antibodies. The topmost band (marked by a red dot) indicates the target protein in this experiment, while the bottom bands are degradation products.
Toward understanding a functional role for AFF1 acetylation in transcriptional regulation, we first identified AFF1 domains that could be acetylated by p300. Different domains of AFF1 have been shown to interact with different SEC components (14). The C-terminal end of AFF1 interacts with AFF4, whereas the extreme N terminus is involved in the interaction with P-TEFb. The AFF1 transactivation and AF9/ENL interaction domains are closer to the N and C termini, respectively. To map acetylated domains, 293T cells were cotransfected with plasmids expressing p300 and various AFF1 domains. The results show no loss of acetylation with a C-terminal deletion to residue 1,012, but a complete loss of acetylation with a further deletion to residue 812 (Fig. 1E, lanes 15 and 16), thus suggesting acetylation sites within residues 813 to 1,012. However, 2 C-terminal fragments (301 to 1,211 and 701 to 1,211) containing these regions showed only modest acetylation when compared to full-length AFF1 (Fig. 1E, lanes 19 and 20), further suggesting that p300-mediated acetylation within the 813 to 1,012 amino acids also requires an additional N-terminal domain. Since the C terminus of AFF1 also contains its nuclear localization signal, we wondered whether defects in AFF1 acetylation in other fragments might reflect their inability to localize to the nucleus for proper acetylation. Consistent with this hypothesis, a localization study clearly showed that the AFF1 acetylation-defective fragments (1 to 812, 1 to 412, and 1 to 212) are primarily localized in the cytoplasm (Fig. 1F). Since the functional activity of p300 is primarily restricted to the nucleus (32), an inability of the AFF1 fragments to enter nuclei may also explain the absence of acetylation signals in those fragments (Fig. 1E).
Since AFF1 is specifically acetylated by p300 within mammalian cells, we asked whether AFF1 could interact with p300. Cotransfection of 293T cells with AFF1 and p300 plasmids and subsequent immunoprecipitation analysis showed an interaction between ectopically expressed AFF1 and p300 (Fig. 1G, lane 4). To rule out interaction artifacts arising from overexpression, we immunoprecipitated endogenous AFF1 protein in 293T cells with an AFF1-specific antibody and confirmed by immunoblot an interaction between endogenous AFF1 and p300 (Fig. 1H). To identify the specific AFF1 domain that interacts with p300, 293T cells were cotransfected with AFF1 fragment- and p300-expressing plasmids. Subsequent coimmunoprecipitation and immunoblot analyses showed that, whereas an AFF1 fragment between amino acids 301 and 700 is important for AFF1 interaction with p300 (Fig. 1I, compare lane 9 with lane 10), a fragment (1 to 812) containing this region itself fails to interact with p300 (Fig. 1I, lane 6). These results indicate that, along with region 301 to 700, an additional region beyond residue 812 is also important for AFF1 interaction with p300. Therefore, based on all these results, we conclude that human AFF1 protein interacts with and is acetylated by p300 through a mechanism that involves multiple AFF1 domains.
AFF1 Is Directly Acetylated at Specific Regions by p300 in Vitro.
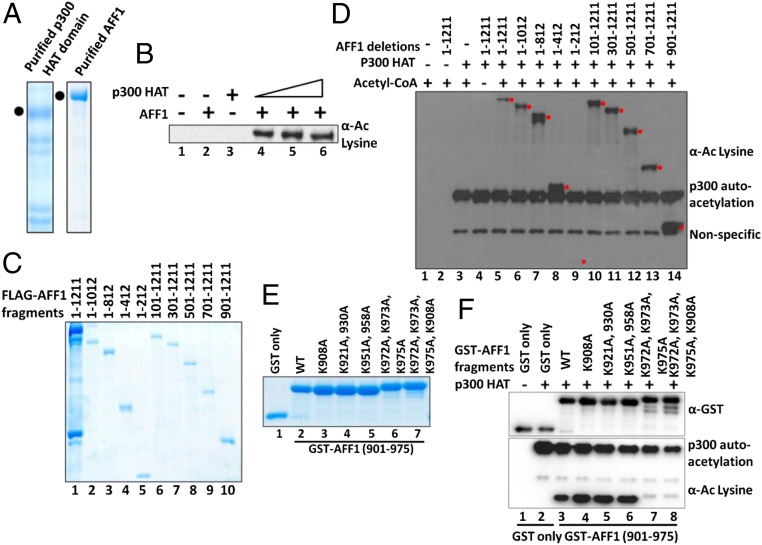
To address whether AFF1 is a direct target for p300-mediated acetylation, we purified AFF1 and the p300 HAT domain following expression from baculovirus and bacterial vectors, respectively (Fig. 2A). An in vitro acetylation assay and subsequent immunoblot with pan-acetyl lysine-specific antibody showed p300-dependent acetylation of AFF1 (Fig. 2B, compare lane 2 with lanes 4 to 6), thus confirming AFF1 as a bona fide target for p300-mediated acetylation. To map the acetylated domains, AFF1 fragments were expressed from baculovirus vectors, purified (Fig. 2C), and tested for acetylation by p300. The results show that, except for the N-terminal 1 to 212 fragment, all other fragments were acetylated by p300. The discrepancy between these results (notably acetylation of the 1 to 812 and 1 to 412 fragments) and those of the cell-based assays indicating C-terminal domain-restricted acetylation could reflect the cytoplasmic localization of corresponding nonacetylated fragments in the cell-based assays (Fig. 1E). Further, the observed in vitro acetylation of the AFF1 701 to 1,211 fragment (Fig. 2D, lane 13) that shows no interaction with full-length p300 within mammalian cells (Fig. 1I) likely reflects less stringent in vitro conditions and an AFF1 (fragment) interaction with the truncated p300 HAT domain that was used in the assay system (SI Appendix, Fig. S1A). However, the combined in vitro and cell-based assays agree with respect to p300-mediated acetylation of a C-terminal region but not the N-terminal 1 to 212 fragment that interacts with P-TEFb.
Fig. 2.
Site-specific acetylation of AFF1 by p300 in vitro. (A) Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE)–Coomassie staining showing purified AFF1 and p300 HAT domain proteins. AFF1 and p300 proteins were expressed in baculoviral and bacterial systems, respectively, purified, and resolved by 10% SDS/PAGE and stained by Coomassie brilliant blue. (B) Acetylation of purified AFF1 by the p300 HAT domain. Acetylation of AFF1 (in A) was monitored by immunoblot using pan-acetyl lysine-specific antibody. (C) SDS/PAGE–Coomassie staining of purified AFF1 domains. Indicated AFF1 fragments were expressed through baculovirus vectors, purified and analyzed as in A. (D) Acetylation of purified AFF1 domains by the p300 HAT domain. Acetylation of AFF1 domains (in C) was monitored by immunoblot with pan acetyl lysine-specific antibody. The target protein band is indicated by a red dot in this experiment. (E) SDS/PAGE–Coomassie staining of purified AFF1 (901 to 975) fragment and mutant derivatives. Proteins were expressed in bacteria, purified, and analyzed as in A. (F) Acetylation of AFF1 (901 to 975) and mutant derivatives by p300. Acetylation of AFF1 mutants (in E) was monitored by immunoblot using pan acetyl lysine-specific antibody.
To refine the mapping of acetylated domains, smaller bacterially expressed and purified AFF1 fragments (SI Appendix, Fig. S1B) were tested. Although the majority of these fragments were acetylated by p300 in vitro (SI Appendix, Fig. S1C), a shorter fragment containing 901 to 975 amino acids showed higher acetylation relative to most other fragments (SI Appendix, Fig. S1C, lane 10). Importantly, expression of larger fragments containing this region also showed acetylation in 293T cells (Fig. 1E, lanes 19 and 20). Therefore, we conclude that human AFF1 is directly acetylated by p300 in vitro and that, despite broader acetylation events, the region between residues 901 and 975 may potentially contribute more significantly to overall acetylation by p300 both in vitro and within cells. The subsequent focus on this region (below) is justified by the finding of specific acetylated residues critical for function.
Identification of Specific Residues within AFF1 for p300-Mediated Acetylation.
To identify specific amino acids within the 901 to 975 region of AFF1 that are targeted for p300-mediated acetylation, we initially carried out sequence alignments to identify lysine residues that are conserved across multiple species (SI Appendix, Fig. S1D, red letters). AFF1 901 to 975 fragments with lysine to alanine mutations at these sites were expressed, purified (Fig. 2E), and tested for acetylation by p300. The K972A, K973A, and K975A mutations abolished p300-mediated acetylation (Fig. 2F, lanes 7 and 8), and other mutations showed no significant effects on overall acetylation. Thus, we conclude that one or more of the K972, K973, and K975 residues are critical targets for p300-mediated acetylation of the AFF1 fragment (901 to 975) fragment in vitro.
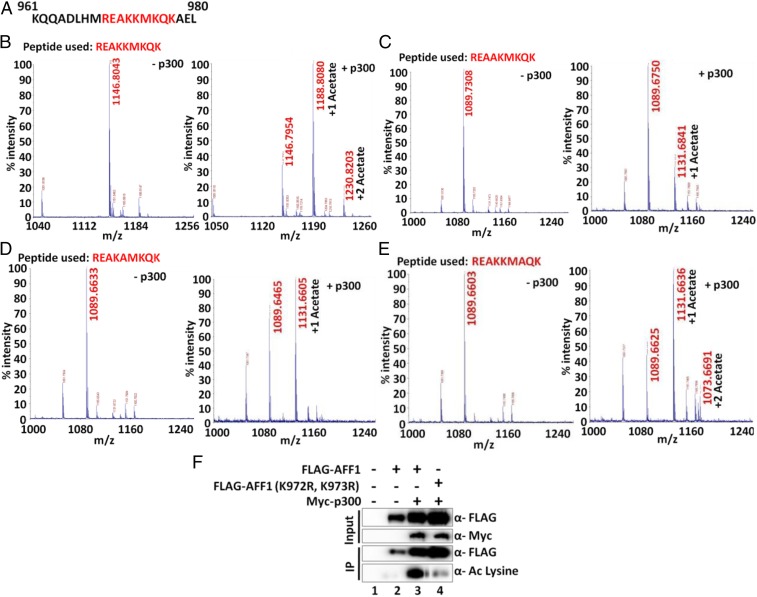
To address the specific nature of these acetylation events, a peptide that spans residues 969 to 977 (Fig. 3A) was subjected to acetylation by p300 and subsequent mass spectrometric analysis. The results showed 2 peaks corresponding to 2 acetylation events in the unmodified peptide (Fig. 3B, compare masses in the presence and absence of p300). Subsequent analyses using peptides containing lysine to alanine mutations showed that, whereas K972A and K973A mutations independently affected dual acetylation events (Fig. 3 C and D), the K975A mutation failed to show any effect on overall acetylation when compared to the WT peptide (Fig. 3E, compare with Fig. 3A). Thus, we conclude that AFF1 K972 and K973 are acetylated by p300 in vitro. Based on these analyses, joint K972R and K973R mutations were introduced into the mammalian AFF1 expression plasmid. Cotransfection of 293T cells with p300 and WT or mutant AFF1 expression plasmids showed significantly reduced acetylation of mutant AFF1 relative to WT AFF1 (Fig. 3F). Therefore, based on our in vitro and cell-based assays, we conclude that human AFF1 is acetylated by p300 in at least 2 conserved lysine residues (K972 and K973).
Fig. 3.
Mass spectrometric analysis of p300-mediated in vitro acetylation of AFF1 using peptides from the 969 to 977 region. (A) Amino acid sequence of AFF1 961 to 980. Peptide sequences used in the in vitro acetylation assay are indicated in red. (B) Mass spectrometric analysis of p300-dependent acetylation of WT AFF1 peptide. Two peaks with masses corresponding to mono- and di-acetylation events are observed. (C) Mass spectrometric analysis of p300-dependent acetylation of mutant (K972A) AFF1 peptide. A single peak with a mass corresponding to a monoacetylation event is observed. (D) Mass spectrometric analysis of p300-dependent acetylation of mutant (K973A) AFF1 peptide. A single peak with a mass corresponding to a monoacetylation event is observed. (E) Mass spectrometric analysis of p300-dependent acetylation of mutant (K975A) AFF1 peptide. Two peaks with masses corresponding to monoacetylation and diacetylation events are observed. (F) Analysis of WT and mutant (K972R, K973R) AFF1 acetylation by p300 in 293T cells. Cells were transfected with indicated plasmids, and anti-FLAG immunoprecipitated samples were analyzed by immunoblot using pan-acetyl lysine-specific antibody.
p300-Mediated Acetylation Reduces AFF1 Interaction with SEC Components within Mammalian Cells.
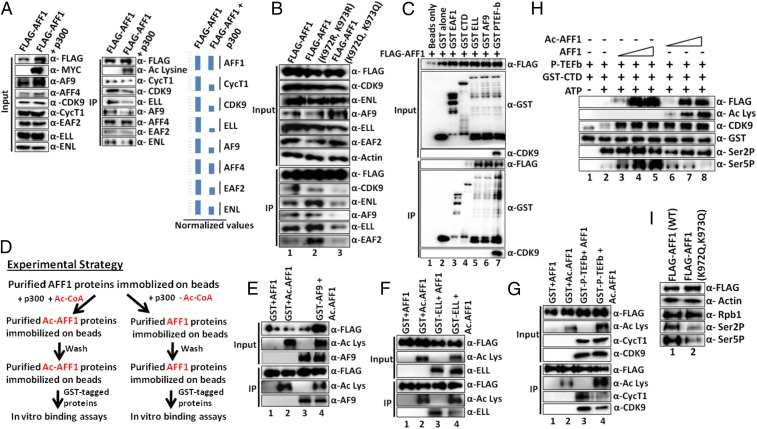
Many transcription factors are known to be functionally modified by acetylation (33). For example, within the SEC components, p300-mediated acetylation of CyclinT1 and CDK9 has been shown to positively regulate transcription, whereas CDK9 acetylation by GCN5 negatively regulates transcriptional activity (29–31). Toward an understanding of a potential role of p300-mediated AFF1 acetylation in transcriptional regulation, we initially tested whether AFF1 acetylation would have any effect on its interaction with other SEC components. To this end, 293T cells were cotransfected with AFF1- and p300-expressing plasmids, and an AFF1 immunoprecipitate was analyzed. The results confirmed the previously observed AFF1 acetylation (above) but also revealed a decrease in acetylated AFF1 interactions with most SEC components relative to control (unacetylated) AFF1 (Fig. 4A, immunoprecipitate immunoblot [Middle] and normalized quantitation [Right]). This reduction is specific, since, in the same assay, we failed to observe a significant effect of acetylation on the AFF1 interaction with its heterodimeric partner AFF4. Next, we asked whether the reduced interaction of AFF1 with other SEC components is indeed a result of site-specific acetylation by p300. We generated 2 AFF1 mutant plasmids in which the target K972 and K973 amino acids were changed either to arginine (K972R and K973R) to maintain charge or to glutamine (K972Q and K973Q) to mimic acetylation. Notably, the arginine mutations did not affect AFF1 interactions with cognate SEC components (Fig. 4B, compare IP blots in lane 2 and lane 1), whereas the glutamine mutations dramatically decreased AFF1 interactions with tested SEC components (lane 3 vs. lane 1). Our results, overall, indicate that p300-mediated site-specific acetylation of AFF1 residues K972 and K973 results in reduced AFF1 interactions with other SEC components, thus suggesting a negative role for p300-mediated AFF1 acetylation in transcription.
Fig. 4.
Site-specific acetylation of AFF1 by p300 reduces its interactions with SEC components both in vivo and in vitro. (A) Effect of p300-mediated AFF1 acetylation on interactions with SEC components. (Left and Middle) The 293T cells were cotransfected with indicated plasmids (top), and anti-FLAG immunoprecipitates were analyzed by immunoblot with antibodies indicated on the right. (Right) Quantitation of factor binding to acetylated AFF1 relative to nonacetylated AFF1. Normalization was done with respect to immunoprecipitated FLAG-AFF1 signal in each lane. (B) Effects of point mutations on intracellular interactions of AFF1 with SEC components. The 293T cells were transfected with indicated plasmids (top) and anti-FLAG immunoprecipitates were analyzed by immunoblot with indicated antibodies (right). (C) In vitro interactions of purified AFF1 and SEC proteins. Immobilized GST-fusion proteins were incubated with purified FLAG-AFF1 (top) and bound proteins were monitored by immunoblot with anti-FLAG antibody (bottom). (D) Experimental strategy for showing a direct effect of AFF1 acetylation by p300 on factor binding in vitro. (E–G) Effects of AFF1 acetylation on AFF1 interactions with SEC components. Analyses were carried out according to the scheme in D and show interactions with (E) AF9, (F) ELL, and (G) P-TEFb. Bound GST-tagged proteins were monitored by immunoblot. (H) Effect of AFF1 acetylation on CTD phosphorylation by P-TEFb. In vitro kinase assays were carried out with the indicated purified recombinant proteins, and reaction products were monitored by immunoblot with antibodies indicated on the right. (I) Effect of overexpression of AFF1 acetylation mimic mutant (K972Q, K973Q) on global Pol II CTD phosphorylation. The 293T cells were transfected with indicated AFF1-expressing plasmids, and Pol II CTD phosphorylation was monitored by immunoblot with indicated antibodies.
p300-Mediated Acetylation Directly Inhibits AFF1 Interaction with SEC Components in Vitro.
Based on the results of our cell-based assays, we next asked whether AFF1 acetylation could directly affect its interaction with cognate partner proteins within the SEC. To first identify SEC components that directly interact with AFF1 in vitro, glutathione S-transferase (GST)-tagged SEC components and FLAG-AFF1 were expressed, purified (SI Appendix, Fig. S2A), and tested for interactions by coimmunoprecipitation. Consistent with our earlier observation of AFF1–partner protein interactions in Sf9 cells (15), our in vitro analyses also showed direct interactions of FLAG-AFF1 with GST-tagged ELL, AF9, and P-TEFb, but not GST alone (Fig. 4C, compare IP blots in lane 2 and lanes 5 to 7). This direct interaction is specific, since, in the same assay, FLAG-AFF1 failed to show any interaction with GST-tagged EAF1 (Fig. 4C, lane 2 vs. other lanes). Consistent with an earlier reported study (34), we also observed a weak interaction between GST-tagged Pol II CTD repeats and AFF1 in this assay (Fig. 4C, lane 2 vs. lane 4).
Based on the direct interaction data above, we next asked whether prior acetylation of AFF1 by p300 would inhibit its interaction with cognate partner proteins in vitro. To address this issue, we followed an experimental strategy (Fig. 4D) in which an immobilized FLAG-tagged AFF1 was acetylated by p300 plus acetyl-CoA, washed extensively to remove any remaining p300, and incubated with purified GST-tagged proteins. Control assays were the same except for omission of acetyl-CoA. As shown in Fig. 4 E–G (lanes 1 and 3 vs. lanes 2 and 4), AFF1 acetylation was readily observed in the presence, but not the absence, of added acetyl-CoA. Notably, whereas the nonacetylated AFF1 retained interactions with AF9, ELL, and P-TEFb complex (lane 3), acetylated AFF1 showed significant reductions in interactions with ELL and P-TEFb complex along with a modest reduction in the AF9 interaction (compare lanes 4 and lanes 3). Since we do not yet know whether the immobilized AFF1 is fully acetylated, the partial loss of SEC components with AFF1 could reflect incomplete AFF1 acetylation. These effects are specific, since, in a similar assay, we failed to observe any effect of AFF1 acetylation on its interaction with Pol II CTD repeats (SI Appendix, Fig. S2B). Therefore, based on all of the collective data, we conclude that p300-mediated site-specific acetylation of AFF1 reduces its interaction with SEC components both in vitro and within mammalian cells.
Acetylated AFF1 Directly Inhibits P-TEFb−Mediated Pol II CTD Phosphorylation.
Among all of the SEC components, enzymatic activity is restricted to the P-TEFb complex that phosphorylates the Pol II CTD repeats, as well as DSIF and NELF components, to regulate transcriptional elongation (35). Our previous study showed the formation of a minimal AFF1⋅P-TEFb trimeric complex mediated by a direct interaction between CyclinT1 and AFF1 (15). Based on our demonstration (above) of a reduced P-TEFb interaction upon AFF1 acetylation, we asked whether acetylation of AFF1 would have any effect on P-TEFb−mediated Pol II CTD phosphorylation. Using a purified recombinant GST-CTD repeat substrate and a purified P-TEFb that was reconstituted in Sf9 cells (SI Appendix, Fig. S2A), we observed that recombinant AFF1 dramatically increased P-TEFb−mediated CTD Ser5 phosphorylation, but not CTD Ser2 phosphorylation (Fig. 4H, lane 2 vs. lanes 3 to 5). Notably, the acetylation of AFF1 completely abolished its stimulatory effect on Ser5 phosphorylation by P-TEFb. We also observed a modest but reproducible inhibitory effect of acetylated AFF1 on P-TEFb−mediated Ser2 phosphorylation. Since autophosphorylation of CDK9 at Thr186 residue in the T-loop region is important for P-TEFb activity (36), we asked whether the reduction of P-TEFb kinase activity by acetylated AFF1 is an indirect effect of CDK9 autophosphorylation. However, as shown in SI Appendix, Fig. S2C, acetylated AFF1 did not significantly affect Thr186 autophosphorylation as monitored by a Thr186 phosphorylation-specific antibody. Based on these observations, we asked whether, as predicted, overexpression of the acetylation mimic mutant of AFF1 (K972Q, K973Q) would inhibit global CTD phosphorylation within mammalian cells. Consistent with this hypothesis, we observed a significant reduction of global Pol II Ser2 and Ser5 phosphorylation upon overexpression of the acetylation mimic mutant, but not WT, AFF1 (Fig. 4I and SI Appendix, Fig. S2D). Thus, we conclude that site-specific acetylation of AFF1 by p300 directly inhibits global Pol II CTD phosphorylation at Ser2 and Ser5 residues.
AFF1 Regulates Expression of a Diverse Set of Genes.
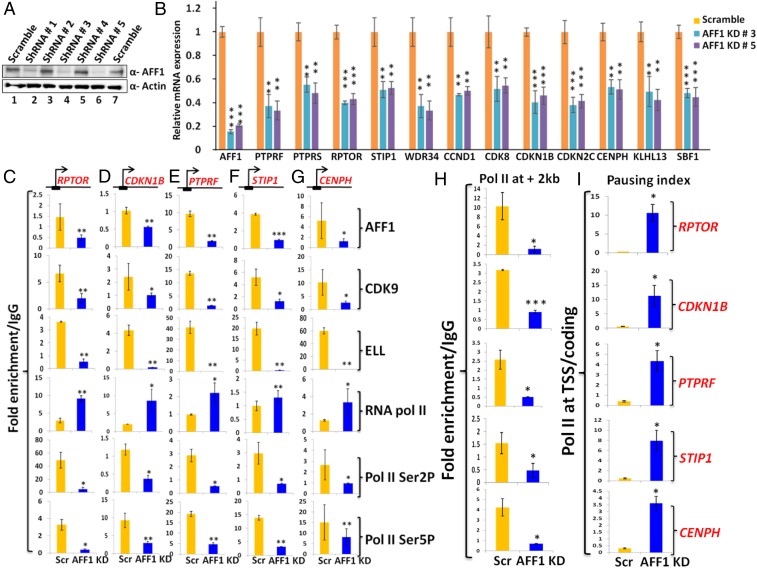
Our findings that p300-mediated AFF1 acetylation results in a reduced AFF1 interaction with SEC components as well as a global reduction of Pol II CTD Ser2 and Ser5 phosphorylation suggested that this acetylation event would directly inhibit transcription of target genes. To address this issue, we initially sought to identify AFF1-regulated target genes and noted an earlier study showing an effect of AFF1 knockdown on global RNA expression in HeLa cells (34). To determine whether similar targets would also be affected in our experimental setup, we generated lentiviruses that stably knock down AFF1 expression in 293T cells (Fig. 5A). Using 2 of these knockdowns, our RNA analyses showed reduced expression of several of the previously identified (34) target genes that were tested (Fig. 5B). The lack of an effect of AFF1 depletion on expression of interacting partner proteins in the SEC (SI Appendix, Fig. S3A) rules out an indirect effect of AFF1 knockdown through reduced expression of these components. Interestingly, the majority of the tested AFF1 target genes that were affected, including CDKN1B, CCND1, CDK8, and CDKN2C, are involved in regulation of the cell cycle and cell proliferation. Consistent with these results, AFF1 knockdown cells showed reduced proliferation and colony formation ability as shown in SI Appendix, Fig. S3 B and C, respectively.
Fig. 5.
AFF1 regulates expression of a diverse set of genes. (A) shRNA-mediated knockdown of AFF1 protein in 293T cells. Cells were transfected with indicated shRNAs, and AFF1 was monitored by immunoblot. (B) Effect of AFF1 knockdown by 2 different shRNAs on gene expression. The 293T cells were transfected with indicated shRNAs, and expression of indicated target genes was monitored by RT-qPCR. The significance of differences in expression is shown for knockdown cells relative to control scramble cells. (C–G) Effect of AFF1 knockdown on recruitment of target factors to TSS regions. The 293T cells were transfected with indicated shRNAs (bottom), and recruitment of indicated factors (right) to indicated target genes (top) was monitored by ChIP-qPCR. The significance of differences in recruitment is shown for knockdown cells relative to control scramble cells. (H) Effect of AFF1 knockdown on Pol II accumulation within coding regions. Pol II association (∼2 kb downstream of TSS) was monitored by ChIP-qPCR analyses of selected genes (indicated on right side of I).The significances of differences in Pol II association are shown for knockdown cells relative to control scramble cells. (I) Pausing indices of Pol II (ratio of total Pol II at TSS/coding region) for indicated genes (right) in control (scramble) versus AFF1 knockdown cells. In all experiments, error bar represents mean ± SD, and statistical analyses were performed using one-tailed Student’s t test wherein * denotes P ≤ 0.05, ** denotes P ≤ 0.01, *** denotes P ≤ 0.001.
To further understand the role of AFF1 in the regulation of target gene expression, and to identify direct target genes, we performed ChIP analysis for SEC components on a few of the target genes that showed reduced RNA expression upon AFF1 knockdown. As expected for direct target genes, AFF1 knockdown resulted in reduced AFF1 recruitment to these genes (Fig. 5 C–G, row 1). Interestingly, and consistent with a role for AFF1 as a scaffolding protein for SEC assembly, a reduced recruitment of CDK9 and ELL at the TSS regions of the tested genes was also observed (Fig. 5 C–G, rows 2 and 3). Consistent with a role for P-TEFb recruitment in the release of promoter-proximal paused Pol II (37), the analyses also showed significant increases in Pol II at the TSS regions of target genes upon AFF1 knockdown (Fig. 5 C–G, row 4). However, in the same ChIP analyses, we observed significant reductions in the CTD Ser2 and Ser5 phosphorylated forms of Pol II at the TSS regions of target genes (Fig. 5 C–G, rows 5 and 6). Since CTD Ser2 and Ser5 phosphorylated forms of Pol II are a reflection of active transcription, a reduction of this signature is another indication of paused Pol II. Consistent with this interpretation, our ChIP analyses further showed reduced levels of Pol II within the coding regions of the tested genes upon AFF1 knockdown (Fig. 5H). In fact, pausing index analyses (total amount of Pol II present at the TSS/coding region) clearly indicated strong increases of Pol II pausing upon AFF1 knockdown (Fig. 5I). Therefore, based on these data, we conclude that human AFF1 regulates the recruitment of SEC components, including P-TEFb, for the release of paused Pol II from TSS regions for transcriptional activation. AFF1 knockdown thus leads to reduced recruitment of SEC components, including P-TEFb, and increases pausing at TSS regions. These results in 293T cells are in general agreement with published results for other target genes in other cell types (34) and set the stage for analysis of effects of AFF1 acetylation (below).
Site-specific AFF1 Acetylation Directly Inhibits SEC Component Recruitment and Target Gene Activation.
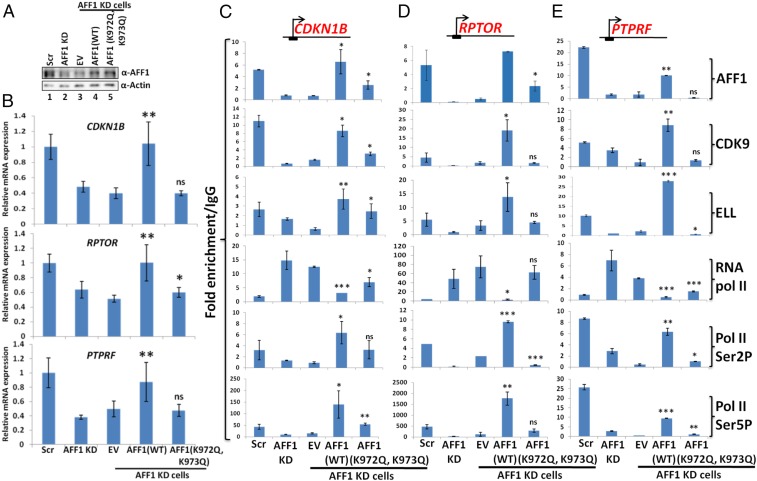
After identifying AFF1 target genes that showed AFF1-dependent SEC recruitment and transcriptional activation in 293T cells, we assessed the effect of AFF1 acetylation on overall factor recruitment and target gene expression. In initial analyses, we restored expression of AFF1 (both WT and the K972Q, K973Q acetylation mimic mutant) in AFF1 knockdown cells through their overexpression (Fig. 6A). Subsequent RNA analyses showed restoration of target gene expression upon reexpression of WT AFF1 but not with the control empty vector (EV) (Fig. 6B, compare EV and WT AFF1). Interestingly, although the ectopic AFF1 acetylation mimic mutant was expressed at a level similar to that of ectopic WT AFF1 (Fig. 6A), it failed to restore transcription of tested target genes and thus indicates a negative role for AFF1 acetylation in target gene activation.
Fig. 6.
AFF1 acetylation mimic mutant (K972Q, K973Q) fails to restore transcription of target genes in AFF1 knockdown cells. (A) Restoration of AFF1 expression, monitored by immunoblot, in AFF1 knockdown cells by overexpression of WT and mutant AFF1 proteins. (B) Effect of WT versus acetylation mimic mutant (K972Q, K973Q) AFF1 on expression of indicated target genes, monitored by qRT-PCR, in AFF1 knockdown cells. The significance of differences in expression for WT versus mutant AFF1 in AFF1 knockdown cells is shown relative to control EV. (C–E) Effect of WT versus acetylation mimic mutant (K972Q, K973Q) AFF1 expression on recruitment of target factors, monitored by ChIP-qPCR, to TSS regions of indicated target genes in AFF1 knockdown cells. The significance of differences in factor recruitment by WT versus mutant AFF1 in AFF1 knockdown cells is shown relative to control EV. In all experiments, the error bar represents mean ± SD, and statistical analyses were performed using one-tailed Student’s t test wherein * denotes P ≤ 0.05, ** denotes P ≤ 0.01, *** denotes P ≤ 0.001, and ns denotes not significant.
Since our in vitro and in vivo assays showed reduced interactions of AFF1 with its cognate SEC partners upon acetylation, we asked whether the AFF1 K972Q, K973Q acetylation mimic mutant would similarly reduce recruitment of interacting SEC partners and thus cause reduced expression of target genes. In further analyses with AFF1 knockdown cells, which showed reduced AFF1, CDK9 and ELL recruitment as observed above (Fig. 5 C–G), ChIP analyses showed that, consistent with the RNA analyses of Fig. 6B, re-expression of WT AFF1, but not an EV, restored recruitment of AFF1 and SEC components ELL and CDK9 (Fig. 6 C–E, compare EV and WT AFF1). The restored levels of recruited factors were similar to the levels of recruited factors in the scrambled short hairpin RNA (shRNA) control cells, further indicating a critical role for AFF1 in overall regulation of SEC recruitment at the TSS regions of target genes. Interestingly, the ectopic AFF1-mediated restoration of SEC recruitment in AFF1 knockdown cells also resulted in increased levels of the CTD Ser2 and Ser5 phosphorylated forms of Pol II and a reduced level of paused Pol II at TSS regions (Fig. 6 C–E). Thus, we conclude that the expression of these target genes is critically dependent on AFF1 through its facilitation of the recruitment of SEC components, including P-TEFb, for release of paused Pol II from TSS regions.
In complementary studies that are consistent with, and support, the results of the WT AFF1 interaction and expression analyses, reexpression of the AFF1 acetylation mimic (K972Q, K973Q) mutant in AFF1 knockdown cells failed to restore recruitment of SEC components, exemplified by ELL and CDK9, at the target genes (Fig. 6 C–E, EV vs. AFF1 [K972Q, K973Q]). Expression of the AFF1 acetylation mimic mutant in AFF1 knockdown cells also failed to restore Pol II CTD Ser2 and Ser5 phosphorylation and thus had little effect on release of paused Pol II. Further, consistent with the differential effects of WT AFF1 versus the acetylation mimic mutant on restoration of factor recruitment and target gene expression in AFF1 knockdown cells, we observed restoration of proliferation and colony formation defects of AFF1 knockdown cells upon reexpression of WT AFF1, but not the acetylation mimic mutant (SI Appendix, Fig. S4). Therefore, based on our detailed biochemical and cell-based assays, we conclude that p300-mediated acetylation of 2 key residues (K972 and K973) in AFF1 results in a reduced interaction of AFF1 with other SEC components as well as a reduced phosphorylation of Pol II CTD Ser2 and Ser5 residues, with consequent reductions in target gene transcription.
AFF1 Is Dynamically Acetylated during Genotoxic Stress Response.
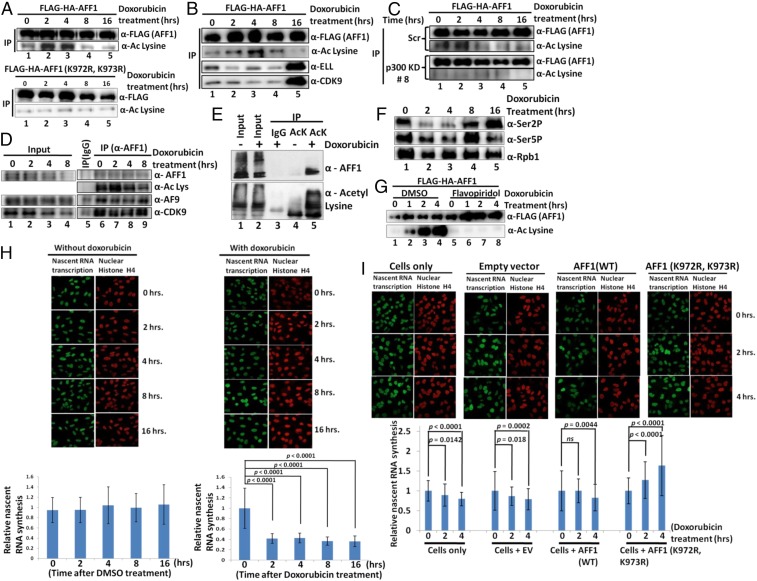
To assess the physiological significance of AFF1 acetylation-mediated transcriptional inhibition in cellular functions, we turned our attention to genotoxic stress-dependent transcriptional repression. Substantial evidence suggests that, upon exposure to genotoxic reagents, mammalian cells initially respond by inhibiting global transcription for proper repair of DNA lesions through efficient recruitment of the repair machinery (1–3, 38). Transcription inhibition also reduces collisions between elongating Pol II and engaged DNA repair machineries. Since acetylation of both histone and nonhistone proteins is involved in dynamic and temporal regulation of functions of transcription factors (28), we asked whether AFF1 acetylation could also play a role in the dynamic regulation of global transcription during exposure to genotoxic stress. We initially tested whether ectopic AFF1 is acetylated in mammalian cells during exposure to genotoxic reagents such as doxorubicin and camptothecin (DNA topoisomerase inhibitors). The 293T cells were transfected with a FLAG-AFF1 expression plasmid and subjected to doxorubicin treatment for different time periods. As shown in Fig. 7A, ectopic FLAG-AFF1 showed increased acetylation as early as 2 h after doxorubicin treatment. This acetylation is dynamic in nature, since the level of acetylation dropped to the normal level by 8 h. The increased AFF1 acetylation is not specific to doxorubicin, since a similar analysis with camptothecin treatment also showed dynamic AFF1 acetylation (SI Appendix, Fig. S5A). Notably, the absence of a similar acetylation signal in a parallel experiment using the AFF1 (K972R, K973R) mutant confirmed K972 and K973 as key targets for this induced acetylation event (Fig. 7 A, Bottom). Consistent with our earlier observation that acetylation of AFF1 reduces its interactions with other SEC components (Fig. 4), we also found that the dynamic AFF1 acetylation was accompanied by concomitant reduction in AFF1 interactions with SEC components ELL and CDK9 (Fig. 7B, lane 1 vs. lanes 2 to 4). As further evidence that the doxorubicin-induced acetylation of AFF1 in cells is mediated by p300, stable shRNA-based p300 knockdown cells (SI Appendix, Fig. S5B) showed significantly reduced acetylation of ectopic FLAG–AFF1 relative to control cells upon doxorubicin treatment (Fig. 7 C, Top vs. Bottom).
Fig. 7.
Transcription-coupled dynamic AFF1 acetylation by p300 inhibits transcription during genotoxic stress. (A) Effect of doxorubicin on dynamic acetylation of WT AFF1 (Top) versus acetylation-defective mutant (K972R, K973R) AFF1 (Bottom). The 293T cells were transfected with indicated plasmids and treated for indicated times. AFF1 acetylation was assessed by immunoblot of anti-FLAG immunoprecipitates using pan-acetyl lysine-specific antibody. (B) Effect of dynamic AFF1 acetylation on AFF1 interactions with SEC components. The 293T cells were transfected with indicated plasmids and treated with doxorubicin for indicated times. Interactions were assessed by immunoblot of anti-FLAG immunoprecipitates with indicated antibodies. (C) Effect of p300 knockdown on doxorubicin-induced dynamic AFF1 acetylation. AFF1 acetylation was monitored by immunoblot in p300 knockdown (Bottom) and control scramble (Top) cells. (D) Effect of doxorubicin on endogenous AFF1 acetylation and factor association. The 293T cells were treated for indicated times, and anti-AFF1 immunoprecipitates were analyzed by immunoblot with indicated antibodies. (E) Effect of doxorubicin on endogenous AFF1 acetylation as evidenced by immunoprecipitation analysis using anti-acetyl lysine-specific antibody. Nuclear extracts of 293T cells treated with or without doxorubicin were immunoprecipitated using pan-acetyl lysine-specific antibody, and immunoprecipitates were analyzed by immunoblot with indicated antibodies. (F) Effect of doxorubicin treatment on dynamic changes in global Pol II CTD Ser2 and Ser5 phosphorylation. The 293T cells were treated for the indicated times, and lysates were analyzed by immunoblot with indicated antibodies. (G) Effect of transcription inhibition on doxorubicin-mediated dynamic AFF1 acetylation. The 293T cells were transfected with a vector expressing FLAG-HA-AFF1 and treated with either DMSO or flavopiridol and doxorubicin as indicated. Anti-FLAG immunoprecipitates were analyzed by immunoblot with indicated antibodies. (H) Effect of doxorubicin on nascent RNA synthesis. HeLa cells were treated for indicated times (Top) and imaged for 5-EU incorporation (Left) or H4 (Right). Quantitated data for nascent RNA synthesis (Bottom) represent averages of 100 cells monitored for each time period. (I) Effect of ectopic expression of WT versus mutant (nonacetylatable) forms of AFF1 on nascent RNA synthesis. HeLa cells were transfected with indicated plasmids and imaged (Top). Quantitated data for nascent RNA synthesis (Bottom) represent averages of 100 cells monitored for each time period. In all experiments, the error bar represents mean ± SD. P values were calculated using one-tailed Student’s t test and ns denotes not significant.
To further validate the above results based on ectopic AFF1 acetylation, we also tested for doxorubicin-induced acetylation of endogenous AFF1. The 293T cells were treated with doxorubicin, and endogenous AFF1 was immunoprecipitated with AFF1-specific antibody. In support of the results with ectopic AFF1, a subsequent immunoblot of the immunoprecipitated AFF1 showed that endogenous AFF1, like ectopic AFF1, is also dynamically acetylated upon exposure to doxorubicin (Fig. 7D, acetyl lysine blot in IP lanes). As observed for ectopic AFF1 acetylation, the dynamic doxorubicin-induced acetylation of endogenous AFF1 also resulted in reduced interaction of AFF1 with SEC components (Fig. 7D, compare lane 6 with lanes 7 and 8 of IP samples). Consistent with an increase in endogenous AFF1 acetylation upon doxorubicin treatment, a further immunoprecipitation analysis using a pan-acetyl lysine-specific antibody showed a significant increase in coimmunoprecipitated AFF1 in nuclear extract from doxorubicin-treated cells relative to nuclear extract from control cells (Fig. 7E, compare lane 5 and lane 6). A subsequent analysis showed a modestly increased p300 interaction with AFF1 around 2 h after doxorubicin treatment (SI Appendix, Fig. S5C). This increased p300 interaction occurs just prior to the time at which maximal AFF1 acetylation is observed, suggesting that the mechanism underlying dynamic AFF1 acetylation may involve a DNA damage-regulated p300−AFF1 interaction. Further, consistent with a role for acetylated AFF1 in reducing global Pol II CTD Ser2 and Ser5 phosphorylation by the P-TEFb component of SEC, we also observed a global reduction of Pol II CTD Ser2 and Ser5 phosphorylation upon doxorubicin treatment around a time window similar to that for AFF1 acetylation (Fig. 7F). Notably, and consistent with earlier reports (5–7), we also observed a reduction in the total level of Pol II at different time points upon doxorubicin treatment (Fig. 7F). However, upon normalization to total Pol II, we observed a stronger correlation of reduced Ser2 phosphorylation, relative to reduced Ser5 phosphorylation, with acetylation of AFF1 (Fig. 7F and SI Appendix, Fig. S5D). Therefore, based on all of the analyses in Fig. 7 A–F, we conclude that human AFF1 is dynamically acetylated within mammalian cells upon exposure to genotoxic reagents and that this acetylation reduces AFF1 interactions with other SEC components and, thus, could potentially play a role in transcriptional regulation.
Dynamic AFF1 Acetylation Is Required for Transcriptional Inhibition and Cell Survival upon Exposure to Genotoxic Reagents.
Toward assessing whether dynamic AFF1 acetylation has any role in regulating transcription, we initially tested whether doxorubicin-induced AFF1 acetylation is dependent on ongoing transcription. To this end, 293T cells were treated with flavopiridol (P-TEFb inhibitor) to inhibit transcription before doxorubicin treatment. Notably, whereas control dimethyl sulfoxide (DMSO) treatment had little effect on dynamic AFF1 acetylation upon doxorubicin treatment, prior flavopiridol treatment completely abolished AFF1 acetylation upon doxorubicin treatment (Fig. 7G, lanes 2 to 4 vs. lanes 6 to 8). Thus, the dynamic doxorubicin-induced AFF1 acetylation is transcription-coupled and requires active transcription. Since the inhibited P-TEFb function is likely at the level of transcription elongation, we hypothesized that dynamic AFF1 acetylation could be an active mechanism of transcriptional inhibition upon sensing of a DNA lesion by the elongating transcription machinery. To address this point, we initially tested the dynamics of transcription in cells after doxorubicin treatment by measuring nascent RNA transcripts through pulse labeling with the uracil analog 5-ethynyl uridine (5-EU) at different time points. Consistent with an earlier report of transcriptional inhibition upon UV treatment (39), we observed a significant down-regulation of nascent RNA transcripts from 2 h onward, whereas control cells showed no reduction in overall transcription (Fig. 7H, compare Left and Right). Notably, the kinetics of transcriptional inhibition is also correlated well with the kinetics of AFF1 acetylation, with maximal transcription inhibition and AFF1 acetylation around similar time points. Interestingly, cells expressing ectopic AFF1 also showed reduced transcriptional activity upon doxorubicin treatment (Fig. 7I, compare cells with AFF1 (WT) and cells with EV. Most interestingly, cells with ectopic expression of the acetylation-defective AFF1 (K972R, K973R) mutant failed to show any reduction in transcriptional inhibition and, in fact, showed a time-dependent increase in nascent RNA transcription (Fig. 7I, compare cells with AFF1 (WT) and AFF1 (K972R, K973R)). The increased transcriptional activity cannot be attributed to differential AFF1 and AFF1 mutant expression, since both were expressed at similar levels (SI Appendix, Fig. S5E). These results argue strongly for a role of site-specific AFF1 acetylation in overall transcriptional inhibition within mammalian cells upon exposure to genotoxic stress. Consistent with a role for p300-mediated AFF1 acetylation in negative regulation of transcription upon exposure to genotoxic stress, p300 knockdown cells (SI Appendix, Fig. S5F) failed to show decreased transcriptional activity (as measured by nascent RNA transcription) at various time points following doxorubicin treatment (SI Appendix, Fig. S5G) when compared to control (scramble shRNA) cells.
Next, we asked whether dynamic AFF1 acetylation-mediated transcriptional inhibition might be a prerequisite for subsequent DNA repair and thus lead to cell survival. We initially tested whether AFF1 knockdown cells showed any reduced colony formation ability after doxorubicin treatment. As shown in SI Appendix, Fig. S5H, AFF1 knockdown results in a dramatic decrease in cell viability after doxorubicin treatment. Consistent with a potential requirement for the reduced transcription for a proper DNA repair response for cell survival, reexpression of WT AFF1 in AFF1 knockdown cells showed increased survival relative to reexpression of acetylation-defective AFF1(K972R, K973R) upon doxorubicin treatment at 2 different concentrations (SI Appendix, Fig. S5I).
Discussion
The major finding of our study is the identification of a dynamic, p300-mediated acetylation of AFF1 that alters SEC assembly and function, resulting in transcription inhibition, during exposure of cells to genotoxic stress. The mechanistic basis for this inhibition relates to the key role of AFF1 as a scaffold for overall assembly of a functional SEC complex with elongation factors P-TEFb, ELL, and other components (13, 15, 24, 25, 27). Consistent with this idea, we have confirmed a direct interaction between AFF1 and other key SEC components. These results necessitate further consideration of AFF1 as a key player in SEC-mediated transcriptional control. The identification of 2 AFF1 lysine residues as essential targets for p300-mediated acetylation, along with the ability of corresponding acetylation mimic mutants to recapitulate the inhibitory functions of AFF1 acetylation, further establishes the potential for AFF1 acetylation to modulate the normal function of SEC in transcriptional activation. Importantly, an increase in cellular AFF1 acetylation upon genotoxic stress, as well as an acetylation-dependent response, further signifies a role for this mechanism in global transcription inhibition during a stress response. A corresponding model is presented in SI Appendix, Fig. S6.
Although multiple mechanisms have been described for transcriptional repression during genotoxic stress response, including an inhibition of transcription initiation (8), a role for elongation factors in the associated dynamic transcriptional regulation has not been appreciated. Since the human SEC plays a major role in controlling transcription elongation (18), dynamic regulation of elongation through posttranslational modification of SEC components is an attractive model for overall transcriptional control during genotoxic stress. Consistent with this idea, a recent study has shown that ATM-mediated phosphorylation of ENL serves to recruit the PRC1 complex for transcriptional repression at DSB sites (23). Our results thus point to another reversible modification (acetylation) of a key SEC component, AFF1, for global transcriptional down-regulation during genotoxic stress. This regulation is clearly dynamic, as evidenced by the reversibility of AFF1 acetylation during early stages of exposure to genotoxic reagents (Fig. 7). Once inhibition of transcription is established through AFF1 acetylation, potentially in collaboration with other mechanisms, this acetylation is decreased. This observation further suggests a possible role for AFF1-mediated SEC assembly for restarting transcription after completion of repair. Indeed, ELL, a known elongation factor within SEC, has been shown to have important roles in transcription restart after UV-induced global DNA DSB repair (22). Notably, the dynamic regulation of AFF1 acetylation during doxorubicin exposure is transcription-dependent, since transcription inhibition by flavopiridol completely blocks AFF1 acetylation. Further, the correlation of AFF1 acetylation with a greater down-regulation of P-TEFb−mediated CTD Ser2 phosphorylation, relative to CTD Ser5 phosphorylation, suggests a direct role for AFF1 acetylation in this regulation.
Our results suggest a role for histone deacetylases in restoring postrepair transcription through AFF1 deacetylation events that could restore AFF1-dependent SEC assembly. As precedent for this idea, deacetylation of SEC component CDK9 by SirT2 and SirT7 has been shown to activate transcription (40, 41). Of note is that a few reports have shown a selective role of Class I HDAC inhibitors in regulating the growth potential of MLL-AF4 cell lines (42, 43). These observations raise the possibility of a role for HDAC1 and HDAC2 in the dynamic AFF1 acetylation associated genotoxic stress response.
How does site-specific AFF1 acetylation affect the overall ability of AFF1 to both interact with SEC components and inhibit P-TEFb−mediated CTD phosphorylation activity? Our observation that the acetylation mimic mutant is able to recapitulate the functions of acetylated WT AFF1 clearly suggests that acetylation alone is sufficient for this regulation. Potentially related is that recent studies suggest that phase separation mechanisms may play important roles in the dynamic regulation of multiprotein associations for biological functions (44–46). Since posttranslational modifications of interacting proteins have been shown to play an important role in the regulation of this differential phase separation (47), it is highly possible that AFF1 proteins may also be subjected to a phase separation mechanism for regulating SEC assembly based on the prediction (IUPred2a [https://iupred2a.elte.hu/]) of significant disordered regions in AFF1 that could enhance phase separation and SEC function (SI Appendix, Fig. S5J).
Although early biochemical and cell-based assays had suggested similar functions for AFF1 and its paralogue AFF4, more recent studies have established that they can be independently incorporated into distinct SEC complexes in cells and may directly regulate expression of distinct sets of genes (34). Our finding that AFF1 is specifically acetylated by p300, whereas AFF4 is not, further supports the notion of nonidentical functions for AFF1- versus AFF4-containing SEC complexes and, especially, differences in their regulation in various cellular processes. It also is notable that, among all SEC components, AFF1 is the most frequent fusion partner of the MLL protein in acute lymphoblastic leukemia and that MLL–AFF4 fusions are rare (48). Since the herein characterized acetylatable lysines in AFF1 are maintained in the leukemogenic MLL–AFF1 fusion proteins, our results raise the possibility of MLL–AFF1 acetylation and either a general role in the various (infant, pediatric, and adult) leukemias or potentially differential acetylation-related functions in these different leukemias. Since the MLL–AFF1 fusion proteins are thought to function through incorporation into corresponding SEC complexes (14), which may be recruited through MLL-related mechanisms, it will be important to determine whether MLL–AFF1 is indeed acetylated in MLL–AFF1 leukemia, whether the N-terminal MLL fragment modulates acetylation of the AFF1 fragment, and whether acetylation, if present, also regulates the function of the MLL–AFF1–SEC complex.
Materials and Methods
Details of materials regarding a list of plasmids, primers for RNA and ChIP analyses, and antibodies used for our study can be found in SI Appendix. Further, methods detailing cell culture techniques, recombinant protein purification, nuclear extract preparation, RNA analysis, ChIP analysis, immunoblot analyses, in vitro interaction analyses, and cell proliferation techniques can also be found in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by a Wellcome-Trust Department of Biotechnology India Alliance Intermediate Fellowship (IA/I/14/1/501287 awarded to D.B.) and National Cancer Institute Grants CA129325 and CA204639 (to R.G.R.). N.K. is a recipient of University Grants Commission Senior Research Fellowship. M.A.H. is a recipient of CSIR Senior Research Fellowship. Generation of 2 important plasmids by Subham Basu and Arijit Nandy is also acknowledged. We also acknowledge Subham Basu and Arijit Nandy for their critical reading and comments on this manuscript. The contribution of CSIR-IICB Matrix-assisted Laser Desorption/Ionization Mass Spec facility for identifying acetylated peptides is also acknowledged. Generous sharing of some plasmids that were used in our studies, through Addgene, by different investigators is also acknowledged.
Footnotes
The authors declare no competing interest.
Data deposition: Raw uncropped images related to this paper have been deposited in the Mendeley Data repository (http://dx.doi.org/10.17632/d7nnkvx87j.1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907097116/-/DCSupplemental.
References
- 1.Adam S., Polo S. E., Blurring the line between the DNA damage response and transcription: The importance of chromatin dynamics. Exp. Cell Res. 329, 148–153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gregersen L. H., Svejstrup J. Q., The cellular response to transcription-blocking DNA damage. Trends Biochem. Sci. 43, 327–341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Svejstrup J. Q., The interface between transcription and mechanisms maintaining genome integrity. Trends Biochem. Sci. 35, 333–338 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Vermeulen W., Fousteri M., Mammalian transcription-coupled excision repair. Cold Spring Harb. Perspect. Biol. 5, a012625 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anindya R., et al. , A ubiquitin-binding domain in Cockayne syndrome B required for transcription-coupled nucleotide excision repair. Mol. Cell 38, 637–648 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson M. D., Harreman M., Svejstrup J. Q., Ubiquitylation and degradation of elongating RNA polymerase II: The last resort. Biochim. Biophys. Acta 1829, 151–157 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Wilson M. D., et al. , Proteasome-mediated processing of Def1, a critical step in the cellular response to transcription stress. Cell 154, 983–995 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gyenis A., et al. , UVB induces a genome-wide acting negative regulatory mechanism that operates at the level of transcription initiation in human cells. PLoS Genet. 10, e1004483 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heine G. F., Horwitz A. A., Parvin J. D., Multiple mechanisms contribute to inhibit transcription in response to DNA damage. J. Biol. Chem. 283, 9555–9561 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lavigne M. D., Konstantopoulos D., Ntakou-Zamplara K. Z., Liakos A., Fousteri M., Global unleashing of transcription elongation waves in response to genotoxic stress restricts somatic mutation rate. Nat. Commun. 8, 2076 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williamson L., et al. , UV irradiation induces a non-coding RNA that functionally opposes the protein encoded by the same gene. Cell 168, 843–855.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He N., et al. , HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol. Cell 38, 428–438 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin C., et al. , AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell 37, 429–437 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokoyama A., Lin M., Naresh A., Kitabayashi I., Cleary M. L., A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell 17, 198–212 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biswas D., et al. , Function of leukemogenic mixed lineage leukemia 1 (MLL) fusion proteins through distinct partner protein complexes. Proc. Natl. Acad. Sci. U.S.A. 108, 15751–15756 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krivtsov A. V., Armstrong S. A., MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 7, 823–833 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Muntean A. G., Hess J. L., The pathogenesis of mixed-lineage leukemia. Annu. Rev. Pathol. 7, 283–301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen F. X., Smith E. R., Shilatifard A., Born to run: Control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 19, 464–478 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Luo Z., Lin C., Shilatifard A., The super elongation complex (SEC) family in transcriptional control. Nat. Rev. Mol. Cell Biol. 13, 543–547 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Shilatifard A., Lane W. S., Jackson K. W., Conaway R. C., Conaway J. W., An RNA polymerase II elongation factor encoded by the human ELL gene. Science 271, 1873–1876 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Shilatifard A., Identification and purification of the Holo-ELL complex. Evidence for the presence of ELL-associated proteins that suppress the transcriptional inhibitory activity of ELL. J. Biol. Chem. 273, 11212–11217 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Mourgues S., et al. , ELL, a novel TFIIH partner, is involved in transcription restart after DNA repair. Proc. Natl. Acad. Sci. U.S.A. 110, 17927–17932 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ui A., Nagaura Y., Yasui A., Transcriptional elongation factor ENL phosphorylated by ATM recruits polycomb and switches off transcription for DSB repair. Mol. Cell 58, 468–482 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Schulze-Gahmen U., et al. , The AFF4 scaffold binds human P-TEFb adjacent to HIV Tat. Elife 2, e00327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulze-Gahmen U., Lu H., Zhou Q., Alber T., AFF4 binding to Tat-P-TEFb indirectly stimulates TAR recognition of super elongation complexes at the HIV promoter. Elife 3, e02375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulze-Gahmen U., et al. , Insights into HIV-1 proviral transcription from integrative structure and dynamics of the Tat:AFF4:P-TEFb:TAR complex. Elife 5, e15910 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi S., et al. , Structural basis for ELL2 and AFF4 activation of HIV-1 proviral transcription. Nat. Commun. 8, 14076 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheikh B. N., Akhtar A., The many lives of KATs—Detectors, integrators and modulators of the cellular environment. Nat. Rev. Genet. 20, 7–23 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Fu J., Yoon H. G., Qin J., Wong J., Regulation of P-TEFb elongation complex activity by CDK9 acetylation. Mol. Cell. Biol. 27, 4641–4651 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabò A., Lusic M., Cereseto A., Giacca M., Acetylation of conserved lysines in the catalytic core of cyclin-dependent kinase 9 inhibits kinase activity and regulates transcription. Mol. Cell. Biol. 28, 2201–2212 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho S., et al. , Acetylation of cyclin T1 regulates the equilibrium between active and inactive P-TEFb in cells. EMBO J. 28, 1407–1417 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dutto I., Scalera C., Prosperi E., CREBBP and p300 lysine acetyl transferases in the DNA damage response. Cell. Mol. Life Sci. 75, 1325–1338 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verdin E., Ott M., 50 years of protein acetylation: From gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 16, 258–264 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Lu H., et al. , Gene target specificity of the super elongation complex (SEC) family: How HIV-1 Tat employs selected SEC members to activate viral transcription. Nucleic Acids Res. 43, 5868–5879 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price D. H., P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20, 2629–2634 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen R., Yang Z., Zhou Q., Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J. Biol. Chem. 279, 4153–4160 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Paparidis N. F., Durvale M. C., Canduri F., The emerging picture of CDK9/P-TEFb: More than 20 years of advances since PITALRE. Mol. Biosyst. 13, 246–276 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Lagerwerf S., Vrouwe M. G., Overmeer R. M., Fousteri M. I., Mullenders L. H., DNA damage response and transcription. DNA Repair (Amst.) 10, 743–750 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Adam S., Polo S. E., Almouzni G., Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell 155, 94–106 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Blank M. F., et al. , SIRT7-dependent deacetylation of CDK9 activates RNA polymerase II transcription. Nucleic Acids Res. 45, 2675–2686 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kosciuczuk E. M., et al. , Sirtuin 2-mediated deacetylation of cyclin-dependent kinase 9 promotes STAT1 signaling in type I interferon responses. J. Biol. Chem. 294, 827–837 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmad K., et al. , Inhibition of class I HDACs abrogates the dominant effect of MLL-AF4 by activation of wild-type MLL. Oncogenesis 3, e127 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stubbs M. C., et al. , Selective inhibition of HDAC1 and HDAC2 as a potential therapeutic option for B-ALL. Clin. Cancer Res. 21, 2348–2358 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hnisz D., Shrinivas K., Young R. A., Chakraborty A. K., Sharp P. A., A phase separation model for transcriptional control. Cell 169, 13–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki H. I., Young R. A., Sharp P. A., Super-enhancer-mediated RNA processing revealed by integrative microRNA network analysis. Cell 168, 1000–1014.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sabari B. R., et al. , Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin Y., Brangwynne C. P., Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Meyer C., et al. , The MLL recombinome of acute leukemias in 2017. Leukemia 32, 273–284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.