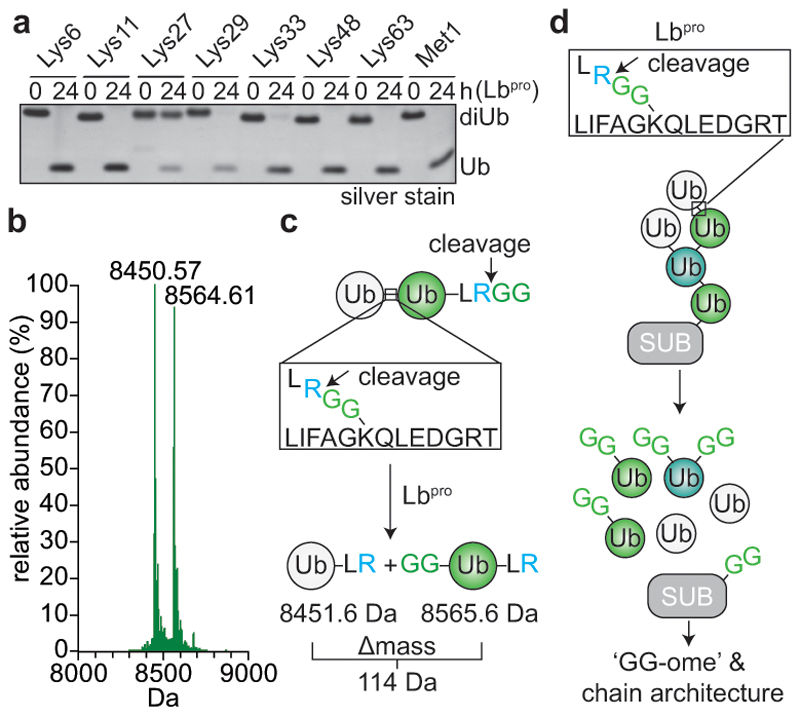
Figure 1. Lbpro cleaves ubiquitin and generates GlyGly-modified proteins.
a, Lbpro was incubated with differently-linked diubiquitin (diUb) for 24 h. Reactions were subjected to SDS-PAGE and visualised by silver staining. A representative example from experiments performed in triplicate is shown. For gel source data, see Supplementary Figure 1. b, Lbpro-treated Lys48-diUb was analysed by electrospray ionization mass spectrometry. Deconvoluted raw spectra are shown (also see Extended Data Fig. 1a). 8450.57 Da peak: distal ubiquitin without the C-terminal Gly75-Gly76 sequence. 8564.61 Da peak (Δmass of 114.04 Da): proximal ubiquitin with apparent wild-type mass; loss of the cleaved C-terminal GlyGly sequence is balanced by GlyGly attachment to Lys48. A representative example of experiments performed in technical triplicate is shown. c, Schematic of Lbpro cleavage sites. Lbpro cleaves diubiquitin twice, after Arg74 of each ubiquitin moiety. d, Lbpro generates a GlyGly-modified proteome, also enabling insights into ubiquitin chain architecture.