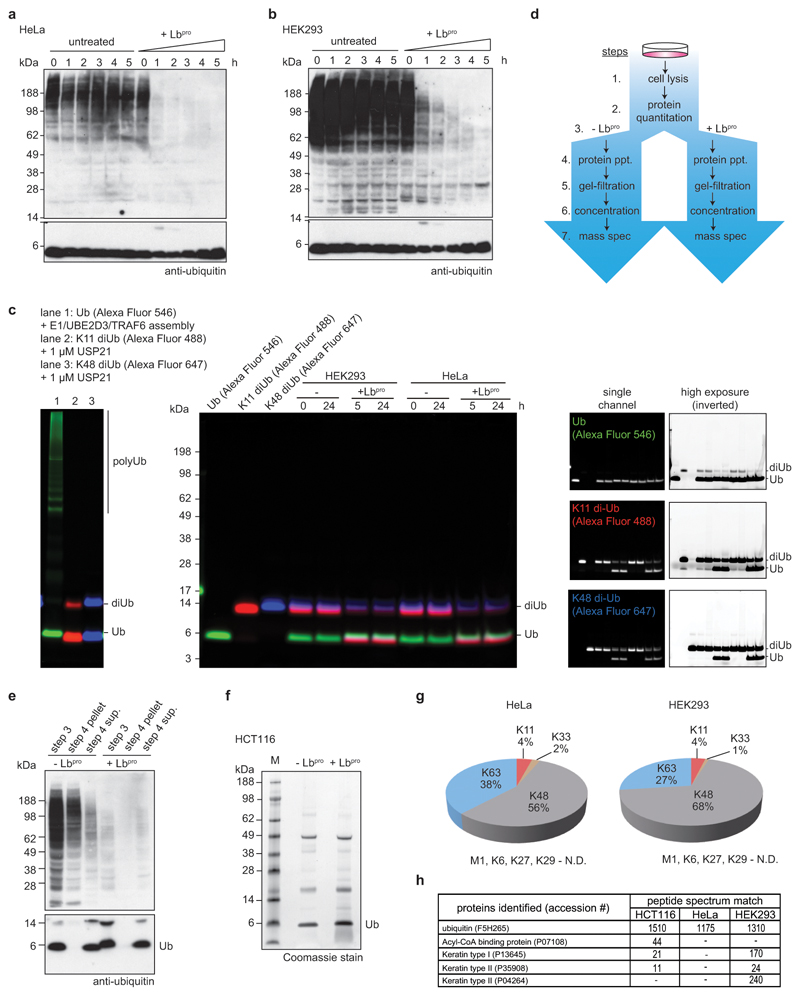
Extended Data Figure 6. Characterisation of Ub-clipping in cells.
a, b Whole cell lysates of (a) HeLa or (b) HEK293 cells were treated with Lbproand analysed by anti-ubiquitin Western blots (compare Fig. 3a). Experiments were performed in triplicate. c, Assessment of endogenous ubiquitin ligase or deubiquitinase activity during Lbpro treatment. HEK293 and HeLa lysates were incubated with fluorescently labelled monoubiquitin, Lys11-linked diubiquitin, and Lys48-linked diubiquitin for the indicated time points. Despite being competent for ligation and susceptible to DUB cleavage (left panel), after 24 h incubation there was no visible DUB or ligase activity, even at high exposures (right panels). This experiment was performed in triplicate. d, Workflow of Lbpro ubiquitin purifications from whole cell lysates. e, Recovery of monoubiquitin after precipitation (dialysis in water) as shown in workflow from d. Western blots were performed on one of three experiments. f, Representative purification of ubiquitin from HCT116 cells. Samples were analysed by intact MS enabled by lack of background protein. These experiments were performed three independent times with similar results. g, Ubiquitin purified from HeLa and HEK293 cells in this manner (see c-e) was analysed by AQUA MS. A representative example of experiments performed in triplicate is shown. Relative amounts of ubiquitin chain linkages are very similar to whole-cell lysate tryptic digests23. N.D., not detected. h, Lbpro-generated monoubiquitin bands purified as in e were excised and analysed using a shotgun proteomics approach to identify contaminant proteins. Experiments were performed in triplicate.