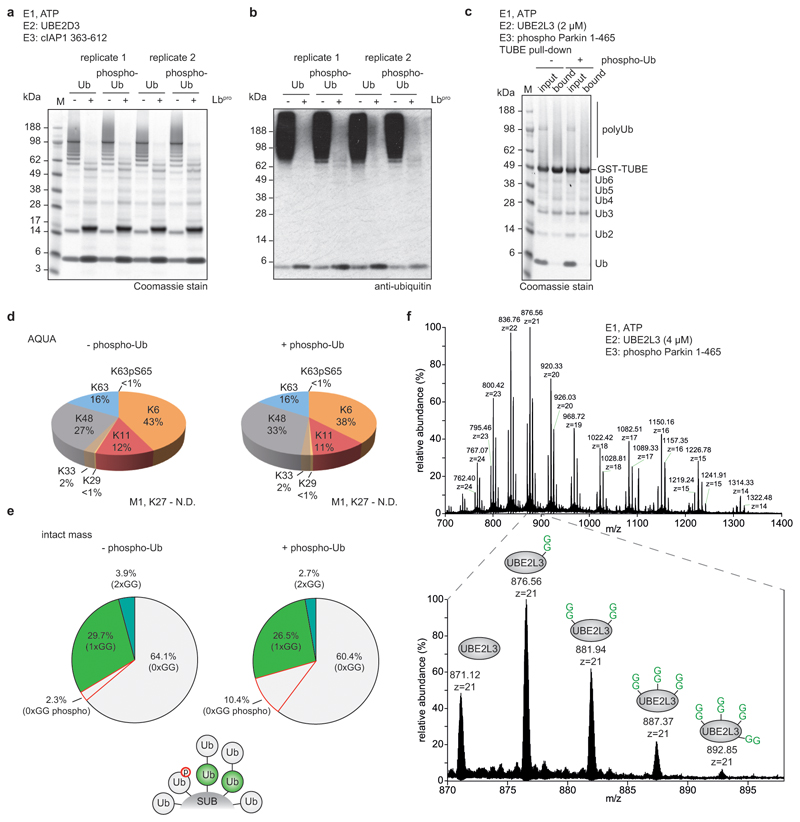
Extended Data Figure 9. Cleavage efficiency of phospho-polyubiquitin chains and MS analysis of Parkin assembly reactions.
a, In vitro assemblies of polyubiquitin from ubiquitin and Ser65 phospho-ubiquitin using cIAP1 and UBE2D3 as previously described11, and their cleavage by Lbpro. Cleavage efficiency was visualised by Coomassie staining. Experiments were independently performed in duplicate with similar results. b, Samples from a visualised by anti-ubiquitin Western blotting. Experiments were independently performed in duplicate with similar results. c, Polyubiquitin assembled by Ser65-phosphorylated human Parkin (pParkin) with UBE2L3 was purified by GST-TUBE pull-downs. Assembly reactions were performed in the absence (-) and presence (+) of 10% total phospho-ubiquitin. Experiments were performed in triplicate. d, TUBE-purified Parkin reactions from c were analysed by AQUA MS to determine chain linkage composition. A representative example of experiments performed in triplicate is shown. e, Lbpro-treated Parkin assembly reactions were analysed by intact MS and spectra deconvolution. A small phospho-ubiquitin contamination is present due to residual PINK1 kinase in the assembly reactions. In these assays, Parkin predominantly assembles monoubiquitin and short chains. Intact MS analysis was performed in duplicate. f, In order to detect a non-ubiquitin substrate with multiple GlyGly modifications by intact mass spectrometry, in vitro assemblies were performed as per e in the absence of phospho-Ub, and with a higher UBE2L3 concentration to facilitate the analysis of UBE2L3. Lbpro-treated assemblies were analysed by LC-MS, revealing up to four GlyGly modifications on a single UBE2L3 molecule. Inset: the distribution of GlyGly-modified UBE2L3 in the +21 charge state. The experiment was performed in triplicate.