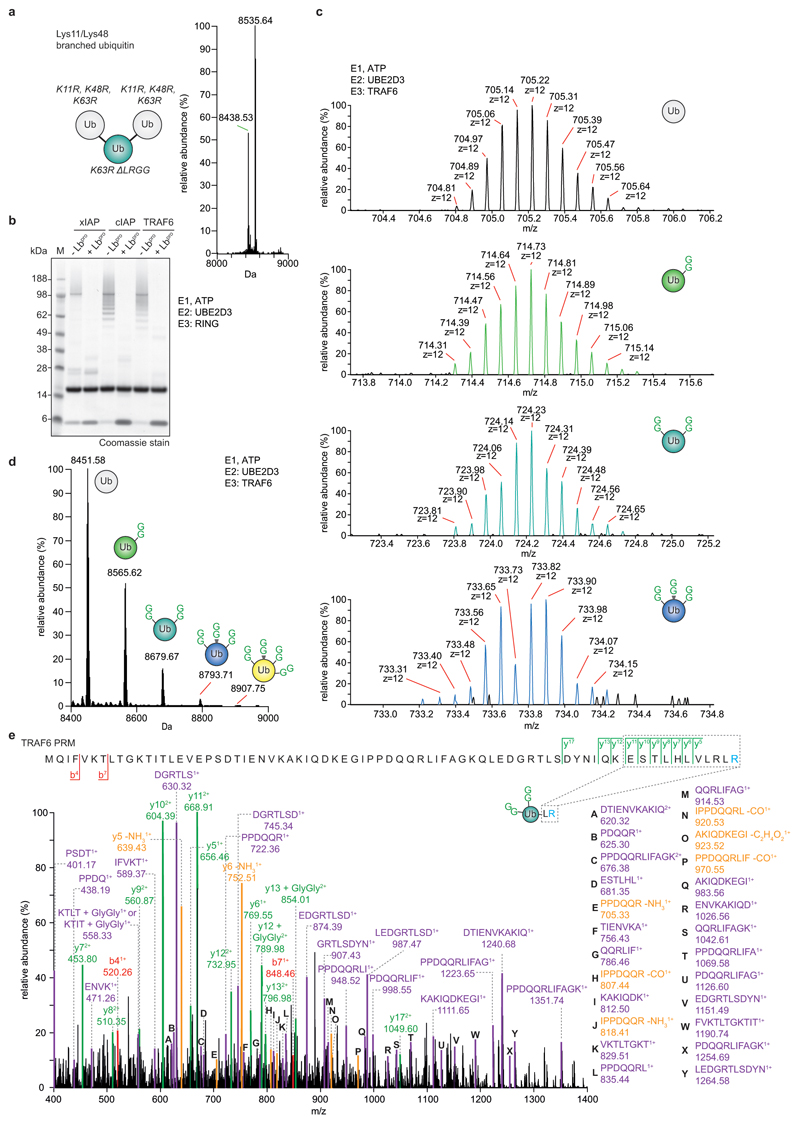
Extended Data Figure 4. Characterisation of branched ubiquitin chains with Ub-clipping.
a, Quantitative intact mass analysis for a branched triubiquitin, in which one ubiquitin moiety is modified on both Lys11 and Lys48 (ref. 49). Lbpro cleavage generates the expected ratio of 1:2 for double-GlyGly and unmodified ubiquitin species. Intact MS analysis was performed in triplicate. b, In vitro assembly reactions from Fig. 2b and 2c. Experiments were performed in triplicate. c, Isotopic distribution of unmodified and different GlyGly-modified ubiquitin species from a Lbpro-treated TRAF6 assembly reaction with UBE2D3 (see b and Fig. 2b). Select spectra are shown from experiments performed in triplicate. d, Intact MS analysis of in vitro TRAF6 assembly reactions from b. Samples were separated by liquid chromatography (LC) prior to MS analysis and spectra deconvolution. LC-MS allowed for the detection of a ubiquitin species with four GlyGly modifications. These experiments were performed three independent times with similar results. e, LC-MS parallel reaction monitoring (PRM) analysis of double-GlyGly-modified ubiquitin as produced by TRAF6 in b, to confirm complete clipping of the C-terminal GlyGly by Lbpro. Green, y ions; red, b ions; purple, internal ions; yellow, neutral loss ions. Experiments were performed in triplicate.