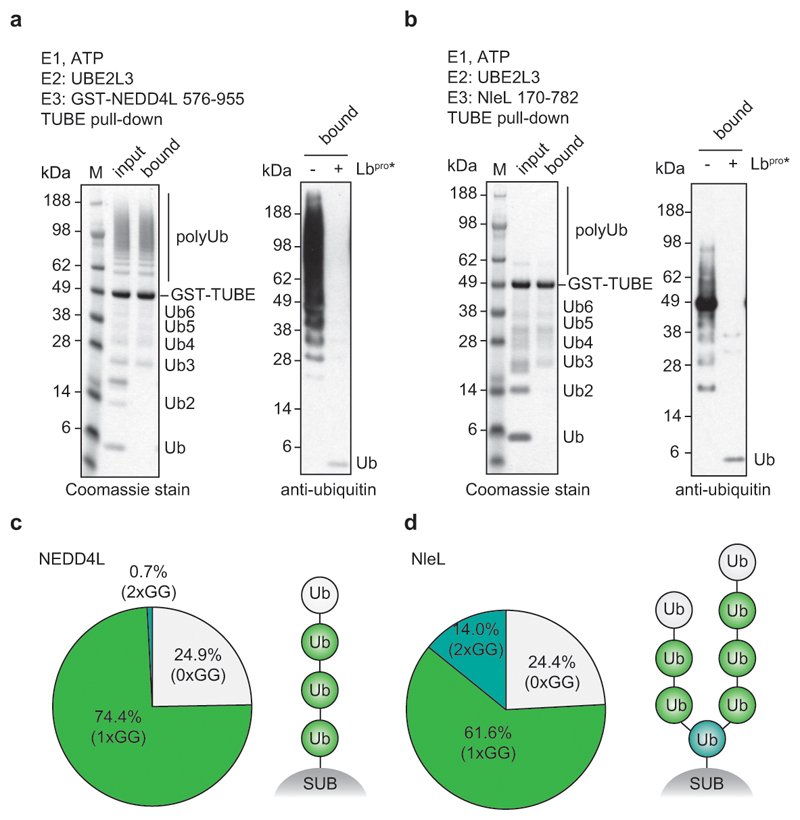
Extended Data Figure 5. TUBE-purified ubiquitin from in vitro assembly reactions.
a, NEDD4L-assembled polyubiquitin chains (see Extended Data Fig. 3e) were purified by GST-TUBE pull-downs (left) and treated with Lbpro(right). TUBE pull-down enriches chains and removes mono- and short polyubiquitin from the reaction (left, bound), and Lbprocleaves TUBE-bound ubiquitin as monitored by anti-ubiquitin Western blots. Experiments were performed in triplicate. b, As in a, using NleL for an E3 ligase known to make branched polyubiquitin39. Coomassie-stained assays were performed in triplicate and Western blots in duplicate. c, Intact MS analysis of samples from a shows relative amounts of each identified ubiquitin species. A ratio of 3:1 for GlyGly-modified vs. unmodified ubiquitin from the NEDD4L-assembled polyubiquitin smear suggests that the average chain length in the reaction is four ubiquitin molecules. d, Samples from b were analysed as in c. A significant fraction of branched ubiquitin suggests that a large percentage of all chains in the reaction are branched. Relative quantities of roughly 2:5:1 for unmodified vs. GlyGly-modified vs. double-GlyGly modified ubiquitin lead to potential ubiquitinated species including the one depicted schematically. The caveat with this model is that chains in the reaction account for a range of lengths. Assays from c and d were performed in duplicate.