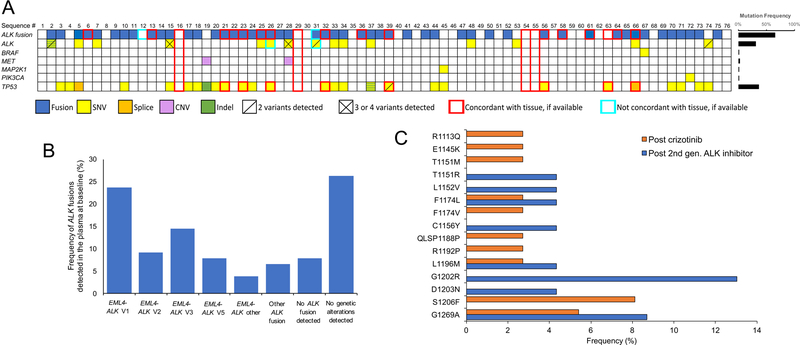
Figure 1. Detection of molecular alterations in plasma from patients with ALK+ NSCLC enrolled in the eXALT2 trial.
(A) Summary of all mutations identified by individual patient at the beginning of treatment with ensartinib. Alterations are color-coded per the figure legend below the image. The red or brown line around a particular mutation (or lack thereof) means that the mutation is either confirmed or not confirmed in the tissue, respectively. The mutation frequencies for each gene are graphed in the right panel, with the denominator equal to total number of patients (i.e., 76). Boxes with yellow and green alternating strips represent a gene for which both an SNV (yellow) and an indel (green) were detected. KRAS, NRAS, NTRK1, RET, and ROS1 were included in the NGS panel; however, no alterations in these genes were detected. ALK, anaplastic lymphoma kinase gene; MET, mesenchymal-epithelial transition gene; MAP2K, mitogen-activated protein kinase 2 gene; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha gene; TP53, tumor protein p53 gene. (B) Frequency of various ALK fusions detected in the plasma at the time of study entrance. “EML4-ALK Other” includes three patients with EML4-ALK fusions that had breakpoints in regions that could not be mapped to a specific variant. An ALK fusion of any kind was not detected in six patients. Note: frequencies in the bar graphs are expressed based upon the total number of patients (n=76). (C) Frequency and distribution of ALK kinase domain mutations detected across the study population. Orange bars represent samples from patients who had received prior crizotinib. Blue bars represent samples from patients who had received crizotinib and at least one second-generation ALK inhibitor. Frequencies are expressed based upon the total number of patients that received post-crizotinib (n=37) or received post-crizotinib and a second-generation ALK inhibitor (n=23).