Fig. 5.
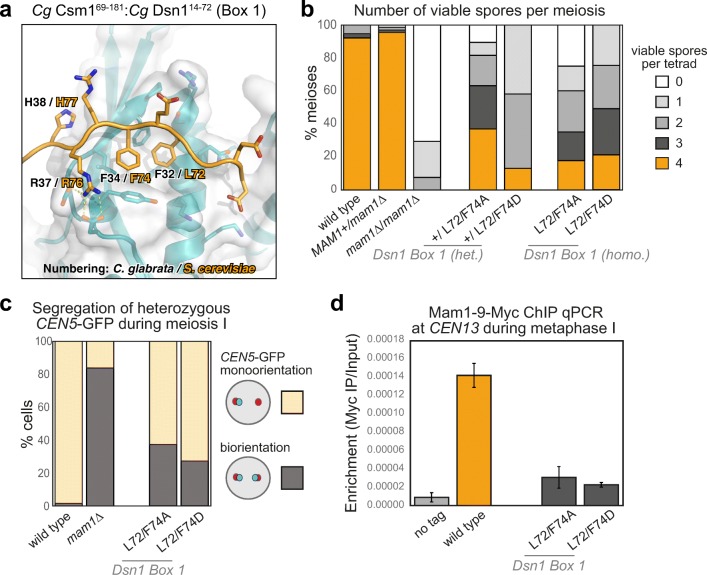
The Csm1-Dsn1 Box 1 interface a Close-up view of the Cg Dsn1 Box 1 region (orange) interacting with the Csm1 conserved hydrophobic cavity (blue with white surface) in the Cg Csm169–181:Cg Dsn114–72 complex. Residue numbers shown are for Cg Dsn1, with Sc Dsn1 equivalents shown in orange text; b Dsn1 Box 1 is critical for meiosis. Spore viability of diploid strains with the indicated genotypes were analysed as described in Fig. 3b. Between 38 and 68 tetrads were dissected for each condition, from a minimum of two independent diploids. Data for wild type and mam1Δ is reproduced from Fig. 3b. Other diploids were generated from matings between AMy1827 and AMy11417 (heterozygous mam1Δ), AMy1827 and AMy17222 (heterozygous DSN1-L72A F74A), AMy1827 and AMy17123 or AMy17313 (heterozygous DSN1-L72D L74D), AMy17222 and AMy17223 (homozygous DSN1-L72A F74A) or AMy17313 and AMy17373 (homozygous DSN1-L72D F72D). c Live cell imaging was used to score sister chromatid co-segregation during anaphase I in cells carrying heterozygous CEN5-GFP foci and Dsn1 Box 1 mutations as described in Fig. 3 c, d. Data for wild type and mam1Δ is reproduced from Fig. 3 d, other strains analysed were AMy25821 (DSN1-L72A F74A; n = 78) and AMy26543 (DSN1-L72D F72D; n = 59); d Mam1 association with a representative centromere in a metaphase I arrest was analysed by ChIP-qPCR as described in Fig. 4d. Data for wild type and no tag is reproduced from Fig. 4d. Other strains used were AMy25618 (DSN1-L72A F74A) and AMy26543 (DSN1-L72D F72D) and the average of 3 or 5 biological replicates, respectively, is shown with standard error bars