Figure 3.
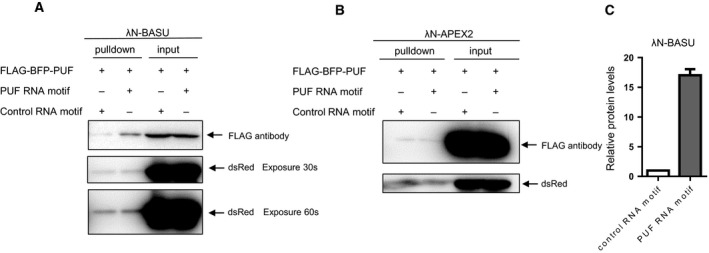
RBPL (λN‐BASU) and RBPL (λN‐APEX2) in RNA–protein interaction detection. (A) Proximity labeling by RBPL (λN‐BASU) on the PUF RNA motif and scrambled control RNA motif. The biotinylation labeling by RBPL showed an excellent signal‐to‐noise ratio via the generation of stable cell lines. (B) Proximity labeling by RBPL (λN‐APEX2) on the PUF RNA motif and scrambled control RNA motif. The RBPL (λN‐APEX2) biotinylation labeling is very weak, with almost no discernible signal between the PUF RNA motif and scrambled controls. (C) Semi‐quantitative analysis of biotinylated PUF proteins by RBPL (λN‐BASU) on the PUF RNA motif and scrambled control RNA motif. The RBPL (λN‐BASU) with PUF RNA motif yielded an approximately 17‐fold enrichment of PUF proteins over scrambled controls. Error bar represents the mean ± SD derived from three independent experiments.
