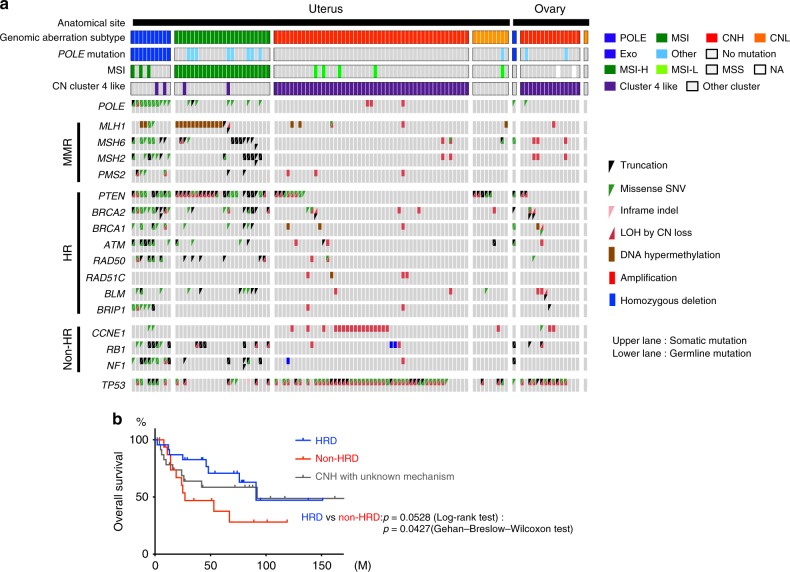
Fig. 2.
Germline and somatic inactivation of DNA repair genes in carcinosarcoma (CS) genomic aberration subtypes. a Relationship between genomic aberration subtype and DNA repair gene mutation. Status of inactivation of POLE, mismatch repair (MMR), and homologous recombination (HR) genes are sorted according to CS subtype and shown along with mutational status of TP53 using Oncoprint. Color code is as follows: truncating mutation, black triangle; missense SNV, green triangle; in-frame indel, pink triangle; loss of heterozygosity (LOH) in wildtype allele by copy-number loss, red triangle; promoter hypermethylation, brown rectangle; amplification, red rectangle; and homozygous deletion, blue rectangle. Somatic and germline mutations are shown in the upper and lower rows, respectively. Using Oncoprint, we show germline missense SNVs only when pathogenic or likely pathogenic, as according to the American College of Medical Genetics and Genomics, Association for Molecular Pathology (ACMG) guidelines (see Methods). Note that 2 of 24 MSI cases (one had MLH1 c.306 + 1_306 + 2delGT, another had MSH2 p.Q374X and MSH6 c.4001 + 2_4001 + 5delTAAC), and 5 of 64 CNH cases (BRCA1 c.5278-1G > A [this case was reported in Abe 201468], BRCA2 p.V1447X, BRCA2 p.T1858fs, BRCA2 p.Y2154fs, and BLM p.K1056fs) had germline mutations, all of which had lost the wildtype allele by LOH or by additional somatic truncation in the tumor samples. b Overall survival of patients with CNH tumors with HRD, non-HRD and the unknown mechanisms. HRD: Homologous recombination deficiency