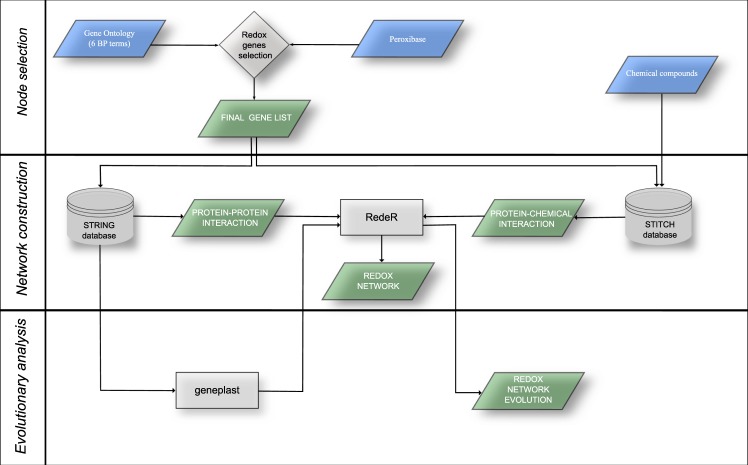
Figure 1.
Analysis workflow. Redox genes were selected from Gene Ontology and PeroxiBase and then manually curated (Node selection). Protein-protein interactions were obtained from STRING database, protein-chemical interactions were obtained from STITCH database, and the final network was handled using RedeR Bioconductor package (Network construction). The evolutionary root of each A. thaliana antioxidant gene was inferred using geneplast Bioconductor package (Evolutionary analysis).